Disclaimer: machine translated by DeepL which may contain errors.
The Rigakubu News
The Rigakubu News, January 2025.
The Frontiers of Research for Undergraduates
Uncovering transcription by removing DNA from histones
Hikaru Nozawa (Second-year student, Master's Program, Department of Biological Sciences)
Sotarou Uemura, Professor, Department of Biological Sciences
DNA does not exist as a long, string-like molecule, but is folded into structures called
nucleosomes, which are stored in the nucleus.
These structures regulate how specific genes are read when needed (transcription).
However, the mechanism of transcriptional activation in
special nucleosomes containing the histone variant H2A.B, which is abundant in testis and cancer cells, was unknown.
In this study, using a novel tool called nanopore measurement, we found that
H2A.B nucleosomes are more unstable and collapse more easily than the normal type, and their pathways are more diverse.
![]()
Nucleosomes are structures around which DNA coils, and eukaryotic genomic DNA is composed of a series of these structures. This structure plays an important role in the storage of genetic information and transcriptional regulation. In particular, nucleosomes in which one of the subunits of the octameric histone protein at the center of the nucleosome is replaced by H2A.B instead of the more common H2A (H2A.B nucleosome) are frequently found in testis and cancer cells and are thought to be involved in transcriptional activation. However, the detailed mechanism of this transcriptional activation has long been unknown.
The transcription process involving RNA polymerase requires that DNA is detached from histones and nucleosomes are disassembled. Therefore, the binding strength between DNA and histones is key in the regulation of transcription. In previous studies, single molecule measurement techniques such as optical tweezers, a technique that uses laser light to capture and manipulate small objects (e.g., cells and molecules) in a non-contact manner, and high-speed AFM, a microscopy technique that enables real-time observation of the dynamic behavior of biomolecules and cell surfaces with high spatial and temporal resolution These methods, however, have been used to measure RNA polymers. However, these techniques could not sufficiently reproduce the situation in which RNA polymerase presses down on DNA and applies tension (Figure). Therefore, we attempted to observe the nucleosome disassembly process in detail using a new measurement technique called solid-state nanopore. Since this technique can reproduce the decay of nucleosomes under conditions similar to those during transcription, it was considered to be a powerful tool for elucidating the structural features of H2A.B nucleosomes (Figure).
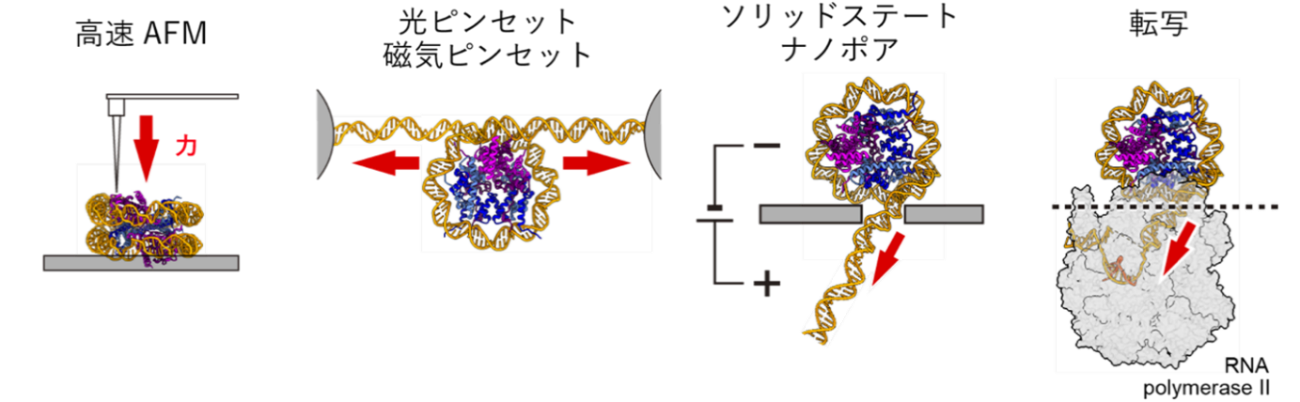
Comparison of conventional and nanopore measurement techniques High-speed AFM applied a force perpendicular to the nucleosome (far left), while optical tweezers applied tension from both sides of the DNA (second from the left). Measurements using solid-state nanopores (second from right) apply tension to the DNA by pressing down on the nucleosome as during transcription (far right).
We first performed experiments in which normal-type H2A nucleosomes and H2A.B nucleosomes were passed through the nanopores, and compared the behavior of DNA under different voltage conditions. The results showed that H2A.B nucleosomes were more likely to disintegrate and dissociate from DNA at lower loadings than the normal type. This indicates that the structure of H2A.B nucleosome is more unstable than that of the normal type.
Furthermore, analysis of the molecular dynamics during passage through the nanopore revealed that some histone dimers in the H2A.B nucleosome rapidly dissociate from the DNA. Molecular dynamics simulations reproduced this disassembly process, suggesting a mechanism for the early dissociation of H2A.B from DNA.
These results suggest that the structural instability of the H2A.B nucleosome allows it to dissociate rapidly from DNA during transcription, thereby facilitating the operation of RNA polymerase. This property may be an important factor contributing to the transcriptional activation of H2A.B.
This study using nanopore measurement opened a new avenue to elucidate nucleosome dynamics during transcription. These results are expected not only to deepen our understanding of the mechanism of transcriptional regulation, but also to lead to future applications in the medical field, such as cancer therapy.
The results of this study were published in H. Nozawa et al. Communications Biology, 7, 1144 (2024).
(Press release on September 20, 2024)


