DATE2024.09.20 #Press Releases
Elucidating the molecular mechanism that pulls DNA wrapped around histones during transcription
Summary
A research group led by Professor Sotaro Uemura, Master Student Hikaru Nozawa, Master Student Satoshi Ogihara, Doctoral Student Rina Hirano, Doctoral Student Munetaka Akatsu, and Assistant Professor Ryo Iizuka at the School of Science, the University of Tokyo, in collaboration with Professor Hitoshi Kurumizaka and Assistant Professor Tomoya Kujirai at the Institute of Quantitative Bioscience, University of Tokyo, Professor Shoji Takada and Doctoral Student Fritz Nagae at the Graduate School of Science, Kyoto University, and Project Lecturer Hirohito Yamazaki at the Nagaoka University of Technology, have demonstrated that solid state nucleosomes containing histone variants H2A.B can normally B nucleosomes are more likely to unwind DNA and dissociate histones in the process compared to the normal type of nucleosome.
Nucleosomes containing the histone mutant H2A.B have been shown to localize in testis and cancer cells, suggesting that they are involved in transcriptional activation. However, the mechanism of their transcriptional activation has not been clarified. In this study, we measured the disassembly process of H2A.B-containing nucleosomes by unwinding DNA in a manner that mimics transcription using nanopore measurement, and found that the structural stability of H2A.B-containing nucleosomes is reduced. Furthermore, we found that there are heterogeneous pathways in the disassembly process. This experimental technique is expected to be important for analyzing the dynamics of nucleosomes during transcription.
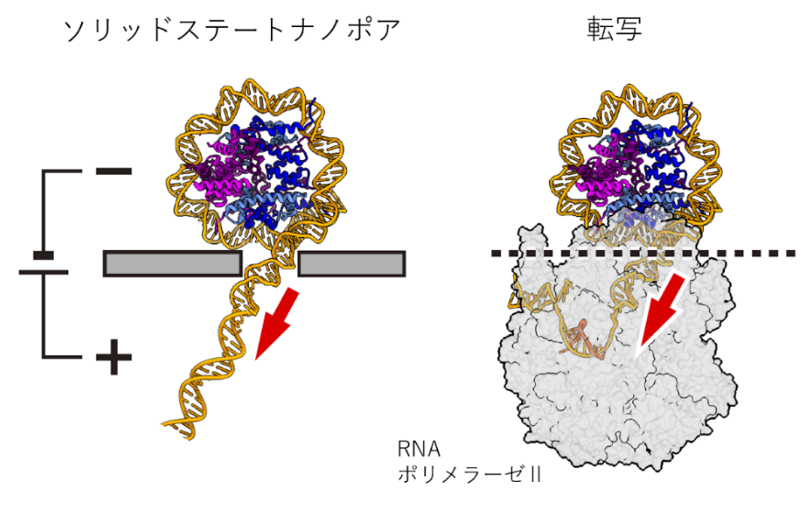
Figure:Conceptual diagram showing forces applied during transfer based on nanopore measurements
Links:Kyoto University, Nagaoka University of Technology, Institute of Quantitative Biosciences, The University of Tokyo
Journal
-
Journal name Communications Biology Title of paper


