DATE2023.01.06 #Press Releases
Ribosome collision-dependent internal cleavage of mRNA
Disclaimer: machine translated by DeepL which may contain errors.
The University of Tokyo Institute of Medical Science
Graduate School of Frontier Sciences, The University of Tokyo
Graduate School of Science, The University of Tokyo
Summary of Presentation
Ribosomes are responsible for the function of protein synthesis by decoding the genetic code on mRNA. Although Cue2 has been identified as an mRNA cleavage enzyme, the mechanism by which Cue2 recognizes ubiquitin modifications and determines the mRNA cleavage site has not been elucidated. Although Cue2 has been identified as an mRNA cleavage enzyme, the mechanism by which Cue2 recognizes ubiquitin modifications and determines the mRNA cleavage site was unknown.
In this study, Professor Toshifumi Inada of the Department of RNA Regulation, Institute of Medical Science, Graduate School of Computational Biology and Medical Sciences, and Department of Information Science, Graduate School of Medical Science, the University of Tokyo, graduate student Shota Tomomatsu of the Graduate School of Pharmaceutical Sciences, the University of Tokyo, and graduate student Atsuya Watanabe of the Graduate School of Pharmaceutical Sciences, Tohoku University, have discovered that the ubiquitin binding activity of endonuclease Cue2 is required for mRNA cleavage on the outer 5' side of the colliding ribosome. They also identified a region at the single amino acid residue level that is essential for mRNA cleavage between colliding ribosomes.
We also analyzed the function of the ribosome-binding molecule Mbf1 in the translation quality control mechanism and found that Mbf1 contributes to the quality control induced by ribosome stagnation and that its function depends on the type of stagnant sequence. These results are expected to lead to a better understanding of mRNA degradation mechanisms and to the development of mRNA vaccines.This research was supported by the Japan Agency for Medical Research and Development (AMED-CREST project number: JP20gm1110010, PI: Toshifumi Inada), Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (project numbers: JP21H05277, JP19H05281, JP22H00401, Toshifumi Inada; 21H00267, 21H 05710, 22H02606, Yoshitaka Matsuo), and Japan Science and Technology Agency (JST) PRESTO (project number: JPMJPR21EE, PI: Yoshitaka Matsuo).
The research results were published in the online edition of the American scientific journal Nucleic Acids Research on December 30.
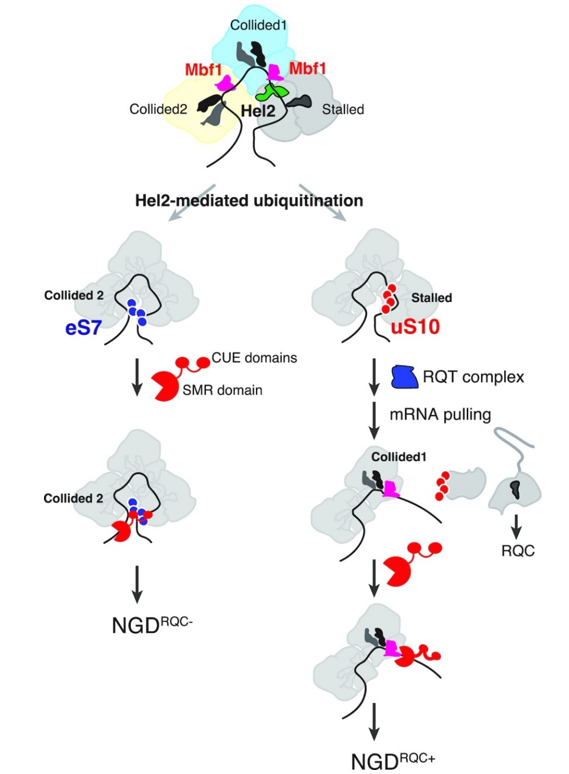
Figure: Model of Cue2 recognition of substrates in two modes of NGD induced by endogenous stagnant sequences; the ubiquitin-binding activity present in the N-terminal region of Cue2 is essential for mRNA cleavage upstream of the colliding ribosome.
Model of NGDRQC- (left): Hel2 forms a K63-binding polyubiquitin chain on eS7 (circled in blue) of the colliding ribosome within the trisome formed on SDD1 mRNA Cue2 binds to K63-binding polyubiquitin chains on eS7 in two CUE domains, CUE-D1 and CUE-D2 binds to the polyubiquitin chain and cleaves mRNA upstream of the colliding ribosome 2.
Model of NGDRQC+ (right): Hel2 ubiquitinates uS10 at the major ribosome and the RQT complex recognizes the polyubiquitinated uS10 (circled in red) The Slh1 helicase subunit of RQT applies tensile force to the mRNA, dissociating the major ribosome into subunits After the mRNA is pulled by the RQT complex, Cue2 cleaves the partially released mRNA from the colliding ribosome.
For more information, please visit the website of the Institute of Medical Science, The University of Tokyo.


