DATE2021.03.02 #Press Releases
Mechanisms that regulate transcription of genes that are densely aligned across the genome.
Disclaimer: machine translated by DeepL which may contain errors.
Soichi Inagaki, Associate Professor, Department of Biological Sciences
Keiyo Oya (2nd Year Doctoral Student, Department of Biological Sciences)
Tetsuhito Kakutani, Professor, Department of Biological Sciences
Key Points of Presentation
- The Arabidopsis thaliana histone demethyltransferase FLD has been shown to regulate transcription by acting in gene regions where transcription occurs in both directions.
- Bidirectional transcription is observed in many genes genome-wide, and this finding may reveal part of the mechanism that facilitates transcription on dense genomes. It was also revealed that this mechanism is used in the mechanism that allows plants to flower at the appropriate time.
- This research has demonstrated a new function of the epigenome (Note 1) in the function of the genome and the regulation of life phenomena, and it is expected that its evolutionary conservation and detailed mechanisms will be elucidated in the future.
Summary of presentation
It is now known that transcription occurs not only in protein-coding genes but also in non-protein-coding regions of the genome, and that transcription occurs in almost all regions of the genome; however, the roles and regulatory mechanisms remain largely unresolved. In particular, the mechanisms that regulate transcription between genes that are close to each other or overlap each other in the genome have not been elucidated.
Associate Professor Soichi Inagaki, graduate student Keiyo Oya, Professor Tetsuhito Kakutani, and their colleagues at the Graduate School of Science, The University of Tokyo, using Arabidopsis thaliana, a plant with a small genome and densely lined genes, have found that transcription in hundreds of gene regions overlaps in the opposite direction, and that transcription in both directions is regulated. We also found a novel chromatin regulatory mechanism that regulates transcription in the regions where this bidirectional transcription occurs. Interestingly, the regulation of genes through this bidirectional transcription is involved in the mechanism by which plants flower in spring after experiencing winter. The results of this study suggest that the relationship of genes to neighboring genes in the genome influences gene regulation, and will hopefully lead to a better understanding of the mechanisms involved.
Announcement
Genes function by being "transcribed" into messenger RNA and "translated" into proteins. analysis. Although some non-coding RNAs have functions in the form of RNAs and are known to play important roles in the regulation of gene expression and genome function, there are also many RNAs whose functions are completely unknown. Thus, it is expected that transcription occurs "densely" in the genome and that there is a mechanism to coordinate transcription between genes (or non-coding RNAs) located close together in the genome, but little is yet understood about this mechanism.
In this study, using the model plant Arabidopsis thaliana, the research group focused on the histone modification (Note 2) localized in the transcribed region of genes and the monomethylation of the fourth lysine residue of the histone H3 protein (with one methyl group attached; called H3K4me1). The research group had previously found a role for H3K4me1 regulation of gene transcription regions in gene activation (Inagaki et al 2017). To further explore the regulatory mechanisms and functions of H3K4me1, they searched for enzymes that exclude H3K4me1 (demethylases). Among them, we focused on one enzyme gene named FLOWERING LOCUS D (FLD), an enzyme gene called lysine-specific demethylase 1 (LSD1), which is widely conserved in eukaryotes from plants to microbes to humans. Using the ChIP-seq method (Note 3), which can examine histone modification patterns throughout the genome, we examined the pattern of H3K4me1 and found regions of higher accumulation of H3K4me1 in the loss-of-function mutants of FLD compared to wild-type plants. On the other hand, the location of FLD protein itself in the genome was also examined by ChIP-seq, and it was located in the region where H3K4me1 accumulates in the mutant. This means that the FLD protein excludes H3K4me1 in these regions and that H3K4me1 accumulates in mutants in which the FLD protein fails to function.
Interestingly, we found that many of the genes in which H3K4me1 accumulates in FLD mutants are convergent genes with the reverse orientation immediately downstream (behind the gene) (Figure 1).
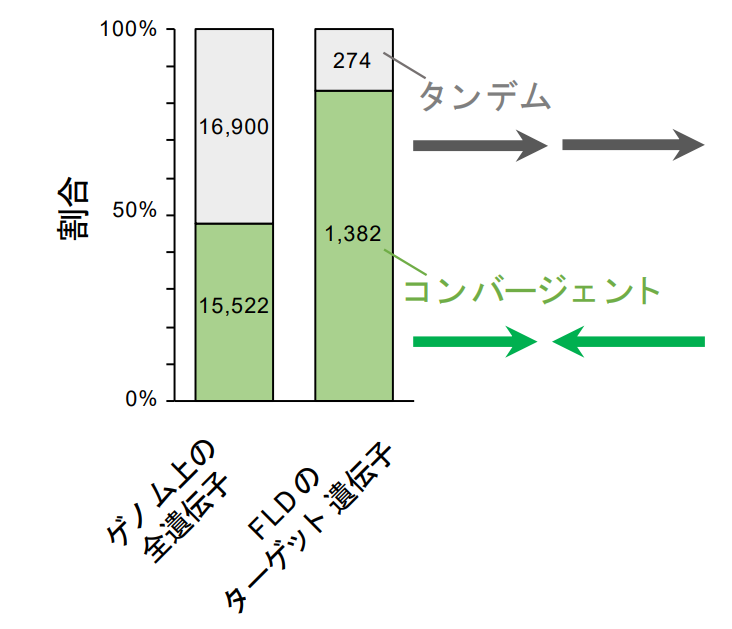
Figure 1: Many of the genes regulated by FLD are convergent genes.
All genes can be divided into "tandem" with the same orientation and "convergent" with the opposite orientation in relation to downstream genes. About half of all genes in the genome are convergent, but most of the genes regulated by FLD are convergent.
Further analysis revealed that in such regions, read-through (Note 4) of transcription from downstream convergent genes occurs, resulting in reverse transcription (antisense transcription (Note 5)). In other words, in addition to normal gene transcription (sense transcription), antisense transcription was also occurring in the region where FLD was working, meaning that transcription was occurring in both directions (Figure 2).
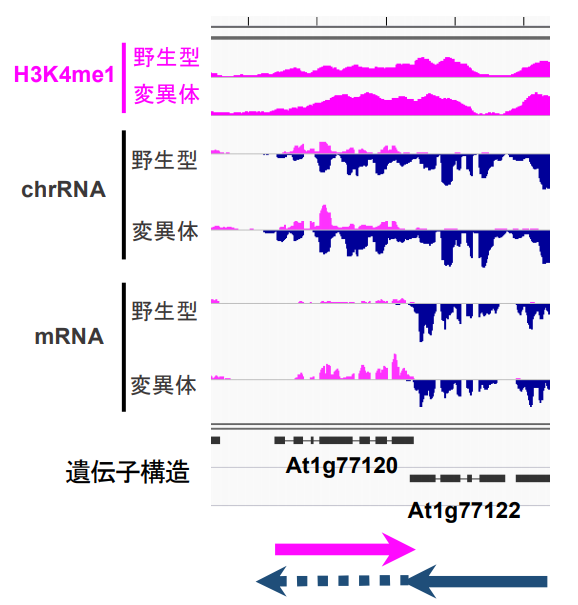
Figure 2: Transcription in both directions is occurring in genes regulated by FLD.
The H3K4me1 at the top is the result of ChIP-seq, and the height of the colored area in the graph represents the amount of H3K4me1. Compared to the wild type, the FLD mutant has increased H3K4me1 in the region of the gene At1g77120. The bottom mRNA represents the expression level of messenger RNA. The pink graph represents transcribed RNA in the rightward direction and the dark blue graph represents transcribed RNA in the leftward direction. The chrRNA in the middle represents RNAs examined by the direct transcription method. chrRNA shows that the At1g77122 gene is transcribed to the region of the At1g77120 gene (represented by the dashed arrow in the figure below), indicating that transcription is occurring in both directions in the At1g77120 gene region.
To explore the significance of FLD excluding H3K4me1 in regions where bi-directional transcription occurs, we analyzed the genome-wide transcription patterns of FLD mutants in detail and found that FLD mutants show increased transcription and faster transcription elongation rate with increased H3K4me1 FLD mutants showed increased transcription and faster transcription elongation rate with increased H3K4me1. It was also found that some of the effects of FLD deficiency on transcription are counteracted by mutants deficient in DNA topoisomerase I, an enzyme that maintains the helical structure of DNA. This suggests that DNA twisting (supercoiling (Note 6) ) due to transcription occurring in both directions is involved in this regulation. Based on these results, we proposed a model in which the regulation of transcription dynamics through the removal of histone modifications by FLD plays a part in preventing collisions and tangling between genes in close proximity on a densely arranged genome (Figure 3).
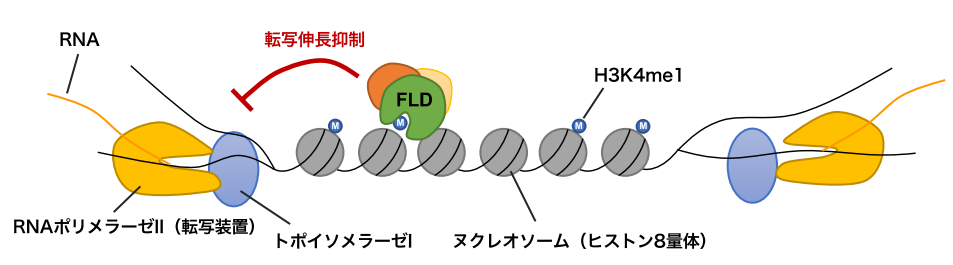
Figure 3: Model of regulation by FLD.
RNA polymerase II proceeds with gene transcription by unwinding DNA double-strands. Topoisomerase I works in concert with the transcription machinery to untwist DNA and help transcription proceed (transcription elongation). Although H3K4me1 is localized in the transcribed region of genes, in the case of genes that are transcribed from both directions, FLD may suppress transcription elongation by excluding H3K4me1, thereby facilitating transcription from both directions.
FLD was also previously known to be involved in the regulation of floral development (Note 7) in plants. Interestingly, FLD was known to regulate floral development by repressing transcription of FLOWRING LOCUS C (FLC), a gene central to floral development regulation, but this study revealed that FLD represses its transcription by excluding H3K4me1 in the FLC region. Since antisense RNAs transcribed in the FLC region play an important role in this regulation, the chromatin regulation mechanism driven by bidirectional transcription found in this study is considered to be "used" in the mechanism that allows plants to flower at the appropriate time (Figure 4).

Figure 4: Regulation of floral development by FLD and topoisomerase.
In mutants deficient in FLD, the FLC gene is no longer repressed and flowering is delayed. Deletion of the topoisomerase I ( TOP1α) gene weakens transcriptional activation of the FLC gene, resulting in earlier flowering in FLD andTOP1α double mutants compared to FLD mutants.
The mechanism for regulating transcription of genes on dense genomes revealed by this study is likely to be conserved in other organisms and may play an important role in the normal function of the genome. This study is also expected to contribute to the understanding of the role of non-coding RNA-mediated transcriptional regulation in the control of life phenomena such as flower formation and the development of artificial control of life phenomena based on this understanding.
This research was supported by the Japan Science and Technology Agency (JST), PRESTO (PI: Soichi Inagaki), and CREST (PI: Tetsuhito Kakutani).
Journals
-
Journal name Nature Plants Title of paper Chromatin-based mechanisms to coordinate convergent overlapping transcription Author(s) Soichi Inagaki*, Mayumi Takahashi, Kazuya Takashima, Satoyo Oya, Tetsuji Kakutani DOI Number 10.1038/s41477-021-00868-3 Abstract URL https://dx.doi.org/10.1038/s41477-021-00868-3
Terminology
Note 1 Epigenome
Genomic information possessed by living organisms is based on the order of DNA bases, but epigenomic information refers to additional information such as DNA methylation and histone modifications, and plays an important role in various life phenomena by influencing the control of gene expression and chromosome behavior. Although more variable than genomic information, it can be inherited across generations and is the basis of epigenetics. ↑up
Note 2 Histone modification
Histones are protein complexes that DNA wraps around in the genome, and their post-translational modifications (methylation, acetylation, phosphorylation, etc.) cause changes in local DNA higher-order structures, etc. It is a typical epigenomic information. ↑up
Note 3 ChIP-seq method
Chromatin Immunoprecipitation (ChIP) is a technique that uses antibodies against specific proteins or protein modifications to recover DNA on the genome where they are present. The recovered DNA can be subjected to large-scale DNA sequencing to determine the location of proteins and their modifications on the genome. ↑up
Note 4 Read-through
When a gene does not terminate transcription at its normal position (read-through of the endpoint), but continues to transcribe. ↑up
Note 5 Antisense transcription
Transcription occurs in the opposite direction of transcription of a protein-coding gene (sense direction). The RNA resulting from this is called antisense RNA. ↑up
Note 6 Supercoil
DNA has a double helical structure, but when the double helical structure is opened during transcription or replication, the twists become stronger in some parts and excessive coiling occurs. DNA topoisomerase is an enzyme that cleaves DNA once and then uncoils it in order to release the supercoiling. ↑up
Note 7: Flowering Regulation
A mechanism that determines when a plant will flower. In Arabidopsis, low winter temperatures epigenetically suppress the expression of the FLC gene, which is responsible for spring flowering. ↑up


