DATE2024.07.11 #Press Releases
Opening the right doors: “jumping gene” control mechanisms revealed
Researchers clarify the molecular mechanisms of chemical mark deposition to prevent “jumping gene” transcription
July 11, 2024
International joint research led by Akihisa Osakabe and Yoshimasa Takizawa of the University of Tokyo has clarified the molecular mechanisms in thale cresses (Arabidopsis thaliana) by which the DDM1 (Decreased in DNA Methylation 1) protein prevents the transcription of “jumping genes.” DDM1 makes “jumping genes” more accessible for transcription-suppressing chemical marks to be deposited. Because a variant of this protein exists in humans, the discovery provides insight into genetic conditions caused by such “jumping gene” mutations. The findings were published in the journal Nature Communications.
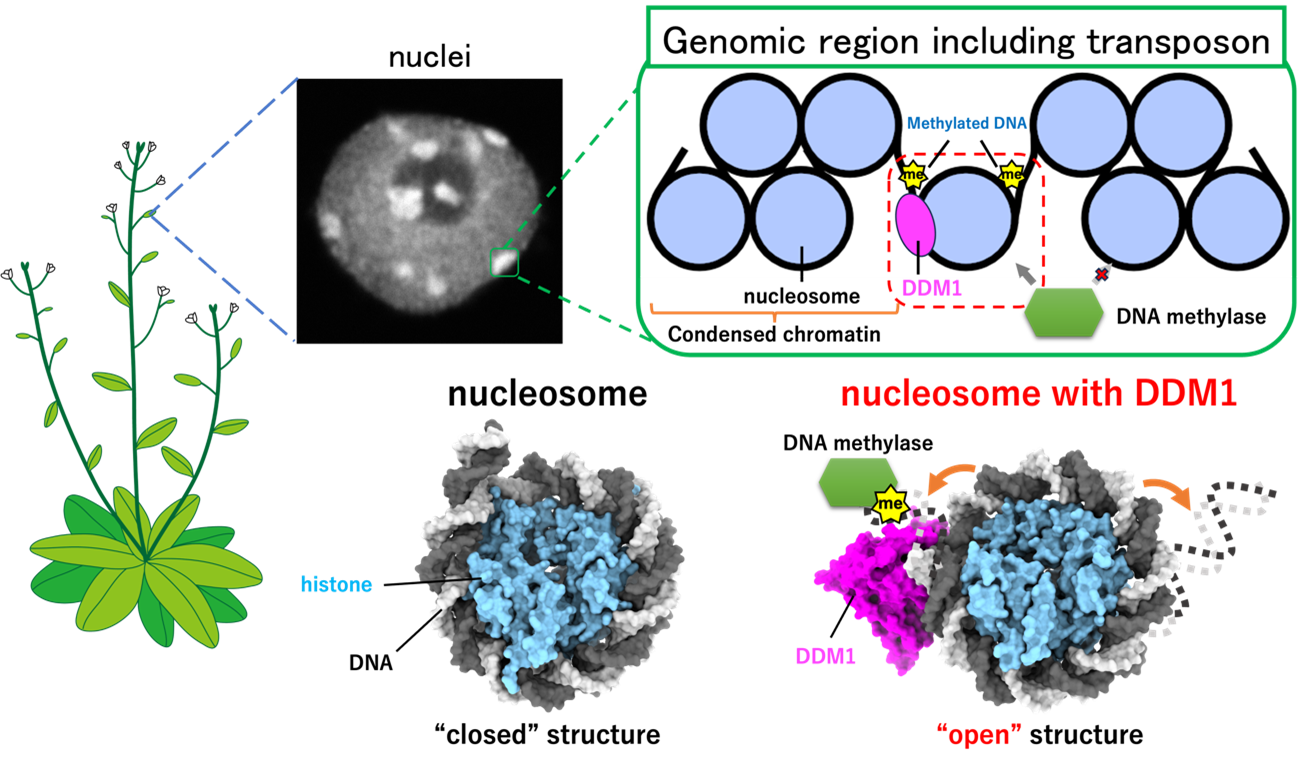
DDM1 protein opening the nucleosome
Disentangled DNA is often referred to as a “string.” In a cell, however, it looks more like a “string ball,” only the looping patterns are much more complex. The smallest unit is called a nucleosome. It consists of a section of DNA wrapped around a protein (histone) scaffolding. Transposons, genes that can “jump” to different locations in the genome, are “tucked away” in nucleosomes, which makes it difficult for the cell to deposit chemical marks that suppress transposon transcription. DDM1 is a protein known for maintaining such suppressing chemical marks, but it has not been clear how it can access transposons when they are neatly “tucked away.”
“Jumping genes are fascinating,” says Osakabe, the first author of the paper, “because they can cause significant changes in the genome, both good and bad. Studying how proteins like DDM1 manage these genes helps us understand the basic mechanisms of life and can have important practical applications.”
The researchers used cryo-electron microscopy, a technique capable of imaging at near-atomic scales. This allowed them to look at the structure of the DDM1 protein and DNA within the nucleosome.
“We felt very excited to see the detailed structures of DDM1 and the nucleosome,” Osakabe recalls. “One of the surprises was how DDM1 opens the nucleosome. It was challenging to capture these structures, but seeing the results made all the hard work worthwhile.”
The high-resolution images showed the exact positions where DDM1 bound to the DNA in the nucleosome. As a result, the specific binding site, which normally closes the nucleosome, got more “flexible” and opened up to allow suppressing chemical marks to be deposited, preventing transposons from being transcribed.
This seemingly minor detail could be the start of major improvements.
“The human version of DDM1, called HELLS, works similarly,” says Osakabe. “In the long term, such discoveries could lead to new treatments for genetic diseases in humans caused by similar genes. This new knowledge also provides insights into how plants and other organisms control their DNA, which could improve our ability to grow better crops or develop new biotechnologies.”
For more details, please read the article:
Akihisa Osakabe, Yoshimasa Takizawa, Naoki Horikoshi, Suguru Hatazawa, Lumi Negishi, Shoko Sato, Frédéric Berger, Tetsuji Kakutani, and Hitoshi Kurumizaka. 2024. Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1. Nature Communications. DOI: 10.1038/s41467-024-49465-w


