DATE2022.09.16 #Press Releases
Identification of a single, stagnant ribosomal sensor factor, Fap1
Disclaimer: machine translated by DeepL which may contain errors.
- Mechanisms to recognize and eliminate loss-of-function ribosomes as abnormal -.
The University of Tokyo Institute of Medical Science
Graduate School of Science, The University of Tokyo
Summary
Ribosomes are responsible for translation activities to decode the genetic code on mRNA and synthesize proteins. Stagnation of ribosomes during translation is a danger signal indicating abnormal protein synthesis, which is recognized and resolved by translation quality control mechanisms. While many quality control mechanisms use collisions between ribosomes as a marker of translation stagnation, the mechanism of translation stagnation resolution targeting a single, stagnant ribosome without collision has been unknown.
In this study, Professor Toshifumi Inada of the Department of RNA Regulation, Institute of Medical Science and Department of Biological Sciences, Graduate School of Science, The University of Tokyo, graduate student Sihan Li of the Graduate School of Pharmaceutical Sciences, Tohoku University, and Professor Roland Beckmann of the Gene Center, University of Munich and his colleagues have identified a novel stagnation sensor factor specific for single ribosomes, the Fap1 marks the single, stagnant, functionally-deficient ribosome and induces its own degradation as well as the resolution of the stagnation.
Comprehensive analysis of ribosome-binding regions on mRNAs by ribosome profiling and structural analysis using cryo-EM revealed that Fap1 preferentially binds to single ribosomes on mRNAs and senses the cessation of movement of mRNAs on both sides. This finding is expected to lead to further understanding of the relationship between ribosome dysfunction and disease development in neurons and other cells in which translation by a single ribosome is active.
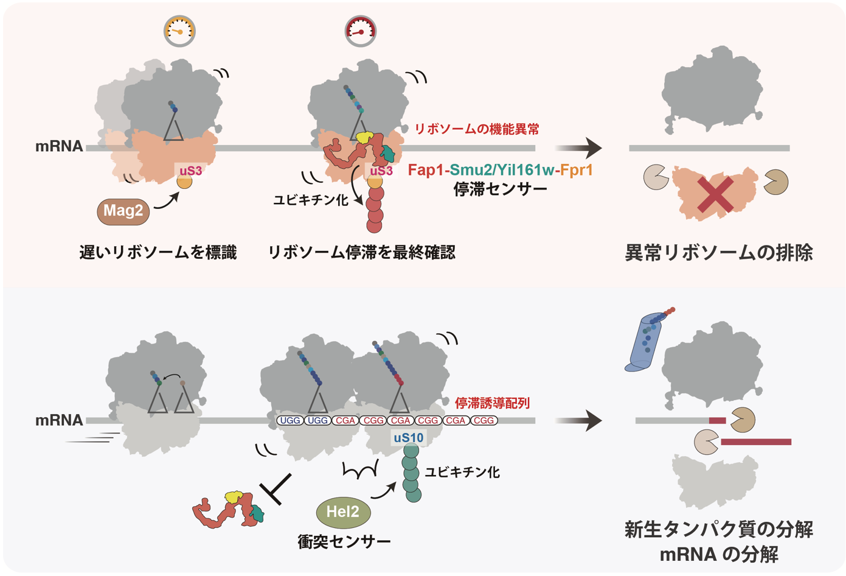
Figure: Comparison between single ribosome stagnation and ribosome collision
Schematic diagram of the pathway leading to the degradation of a single ribosomal stalled ribosome (top). after Mag2 adds the first ubiquitin to uS3 of the ribosome with slow translation rate, when the ribosome is stalled alone, the stasis sensor factor Fap1 binds and elongates the ubiquitin chain. the ubiquitin chain added to uS3 is then translated into the ubiquitin chain of the ribosome. The ubiquitin chain added to uS3 is considered a marker of ribosome dysfunction, causing degradation and elimination of the ribosome itself. On the other hand, when a subsequent ribosome collides with a stagnant ribosome (see below), Fap1 binding is disturbed, activating ubiquitination of uS10 by the collision sensor factor Hel2 and its dependent quality control.
This work was supported by CREST (project number: JP 19gm1110010, PI: Toshifumi Inada), JSPS Grants-in-Aid for Scientific Research (project numbers: JP18H03977, JP19H05281, Toshifumi Inada; 21H00267, 21H05710, 22H02606, Yoshitaka Matsuo), JST PRESTO The research was supported by JST PRESTO (project number: JPMJPR21EE, PI: Yoshitaka Matsuo), JSPS Research Fellowship (project number: 20J20445, Sihan Li), and an Overseas Postdoctoral Fellowship (Ken Ikeuchi).
The research results were published in the online edition of the American scientific journal Molecular Cell on September 15.
For more details, please visit the website of the Institute of Medical Science, The University of Tokyo.


