DATE2022.07.21 #Press Releases
Elucidation of the multimeric structure responsible for the activity of drugs for the treatment of multidrug-resistant bacterial infections.
Disclaimer: machine translated by DeepL which may contain errors.
Takayuki Nakamuro, Project Associate Professor, Department of Chemistry
Eiichi Nakamura, Professor Emeritus Professor, Department of Chemistry, The University of Tokyo
Key Points of the Presentation
- The antibiotic daptomycin is a trump card in the treatment of multidrug-resistant (Note 1) infections. The dynamic behavior of the molecular assembly, which has been assumed to be the key to its activity, and the molecular structure of the tetramer, which is considered to be the active form, have been clarified.
- The molecular hook (Note 2 ), which was specifically designed for fishing daptomycin polymers, was used to capture the dimers, trimers, and tetramers from aqueous solution, and their structures were determined using a state-of-the-art electron microscope with angstrom-millisecond temporal and spatial resolution.
- Methicillin-resistant Staphylococcus aureus has become a major clinical problem. Even the newest drug, daptomycin, has already been found to be resistant to Staphylococcus aureus, and it is expected that a new drug will be developed based on the molecular structure of the multimer revealed in this study.
Summary of presentation
Eiichi Nakamura, Professor, and Takayuki Nakamuro, Project Associate Professor, Department of Chemistry, Graduate School of Science, The University of Tokyo, and their colleagues have clarified the molecular structure of the cyclic tetramer ( LC4 ), which has been assumed to be the key to the biological activity of daptomycin, a cyclic lipopeptide antibacterial drug.
Methicillin-resistant Staphylococcus aureus (MRSA, (*3) ) is responsible for the majority of infections caused by resistant bacteria. Although resistance to daptomycin, the newest MRSA-resistant drug, has already emerged, it is difficult to improve the drug because the mechanism of action at the molecular level is unknown. In this study, daptomycin dimers, trimers, and tetramers were fished out of aqueous solution with a "molecular hook" and their structures were determined using a state-of-the-art high-speed high-resolution electron microscope, (Note 4). The tetramer has a vacancy in the middle of the tetramer, which suggests a mechanism of action such as depolarization by potassium ion efflux through the vacancy. This research group is working to elucidate various chemical phenomena by using a combination of "molecular fish hook" and "high-speed, high-resolution electron microscopy," and is developing a new research field called "imaging molecular science.
The research results were published in the Journal of the American Chemical Society (JACS ).
Publication details
Most resistant bacterial infections in clinical practice are caused by MRSA. Daptomycin (DP, formulation name: Cubicin) has a completely different molecular structure from previous MRSA-resistant drugs, and its unique mechanism of action, calcium-dependent aggregation on the cell surface of Gram-positive bacteria, is thought to be an important process for its activity (Figure 1). The molecular structure of MRSA produced on the cell membrane is not known, and pharmacological studies to create derivatives with higher activity have so far yielded no results (Fig. 2).
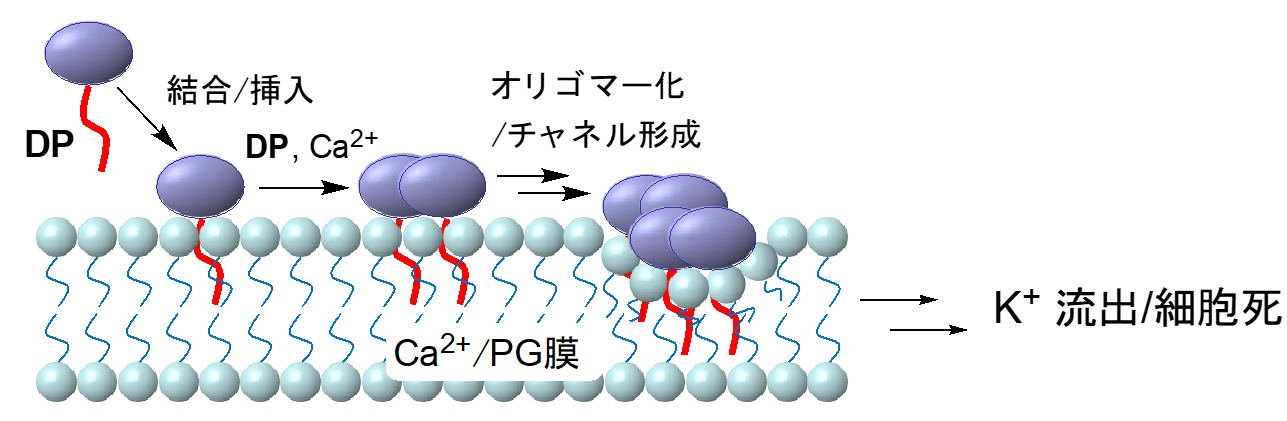
Figure 1: Possible DP mechanism of action. Oligomerization at the cell membrane surface of Gram-positive bacteria is important, and tetramer, octamer, and even larger multimer formation have been proposed.
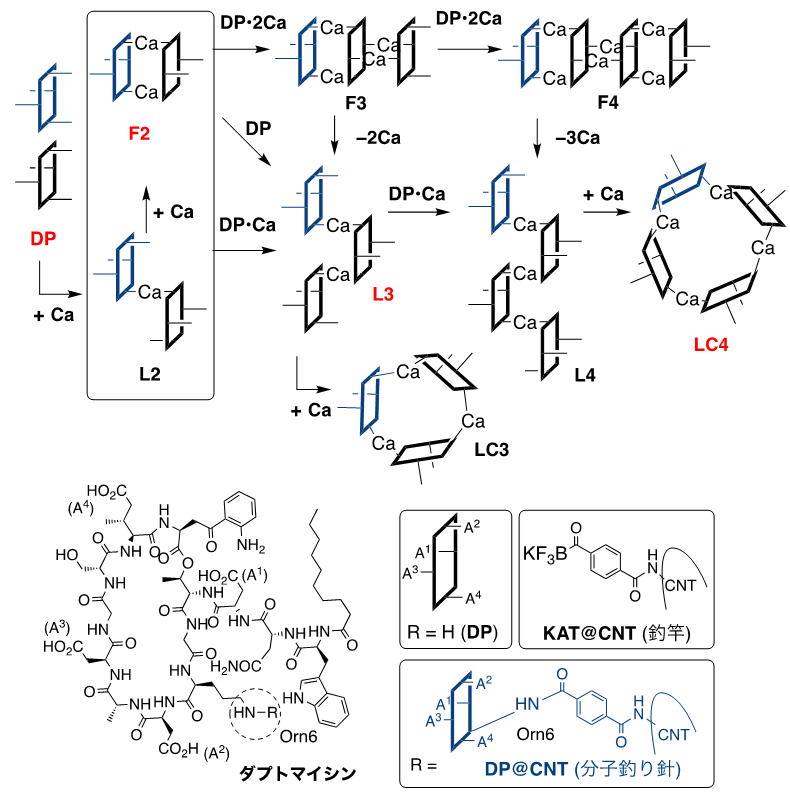
Figure 2: Self-assembly process of DP by calcium ions. The structure shown in red is the molecular species trapped in this study, and the dotted line is ornithine (Orn6) used as the reaction point.
The research group has developed a single-molecule atomic-resolution time-resolved electron microscopy (SMART-EM) method to capture images of the dynamic behavior of various molecules at the atomic level. One method is to chemically modify the tips of carbon nanotubes (CNT (*5) ) to create "molecular hooks" to "fish" for the target molecules. In this study, in collaboration with co-author Professor Jeffrey Bode of the Swiss Federal Institute of Technology Zurich, we developed a method using a single molecule of daptomycin as a "fish hook" in order to clarify the behavior of daptomycin aggregates (Figure 3).
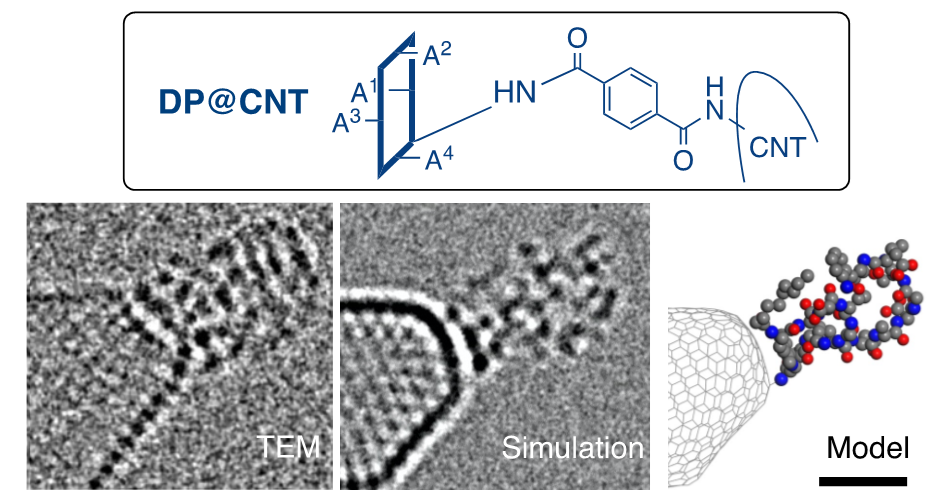
Figure 3: TEM image, simulation, and molecular model of a molecular fish hook, respectively. Scale bar is 1 nanometer.
To investigate the aggregation behavior of daptomycin, we first used dynamic light scattering, a classical analytical technique, to study the dependence of aggregate size in aqueous solution on calcium ion concentration. It was found that the size of the aggregates increased stepwise from monomers to tetramers with increasing concentration. Although this is to be expected, the fact that the dimer is always present regardless of the calcium ion concentration is characteristic, suggesting that the dimer is particularly stable (Fig. 4).
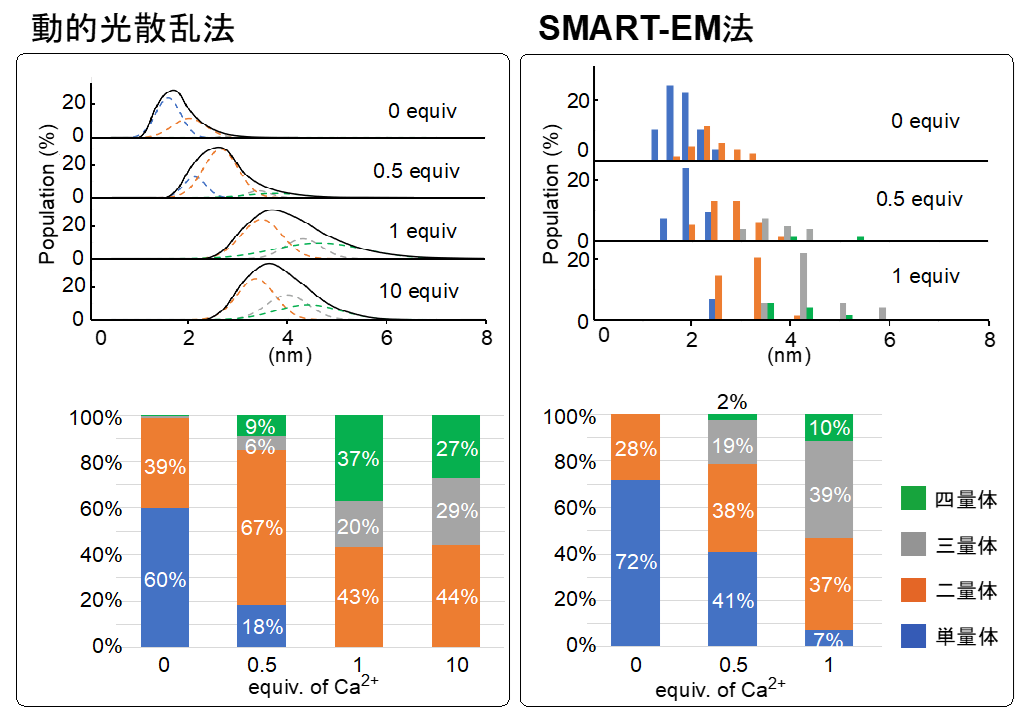
Figure 4: The distribution of molecular species can be estimated by combining dynamic light scattering and SMART-EM.
The fish hooks made in the previous step were then infiltrated with daptomycin and calcium chloride solution. The calcium ions acted as a "glue" to assemble a large number of daptomycin, which allowed us to fish multimeric bodies on top of the CNTs. Specifically, the four carboxylic acid residues (COOH, A1-A4 ) of daptomycin became calcium salts (COO-Ca-OCO) that bonded the molecules together. The structures of about 300 polymers in total were determined by electron microscopy, and the distribution of the polymers in SMART-EM was determined. It is noteworthy that the ratios of polymers determined by dynamic light scattering and electron microscopy were almost identical, and no polymers larger than a pentamer were observed (Fig. 4).
Analysis of the dynamic behavior of the aggregates captured by the fish hook revealed structural fluctuations in the dimer and a linear structure ( L3) in the trimer (Figure 5). As also shown in Figure 2, daptomycin may produce monomeric to tetrameric molecular species in the presence of calcium, and linear and cyclic forms with respect to shape. The molecular images obtained from various angles suggest that the most plausible structure for the tetramer is the cyclic form of LC4. The structure of the daptomycin aggregate, which had not been imagined before, has a vacancy in the center of LC4, suggesting a mechanism of action such as depolarization by potassium ion efflux through this vacancy (Fig. 6).

Figure 5: Molecular images of DP dimer, trimer, and tetramer. See QR code for details. Scale bar is 1 nanometer.
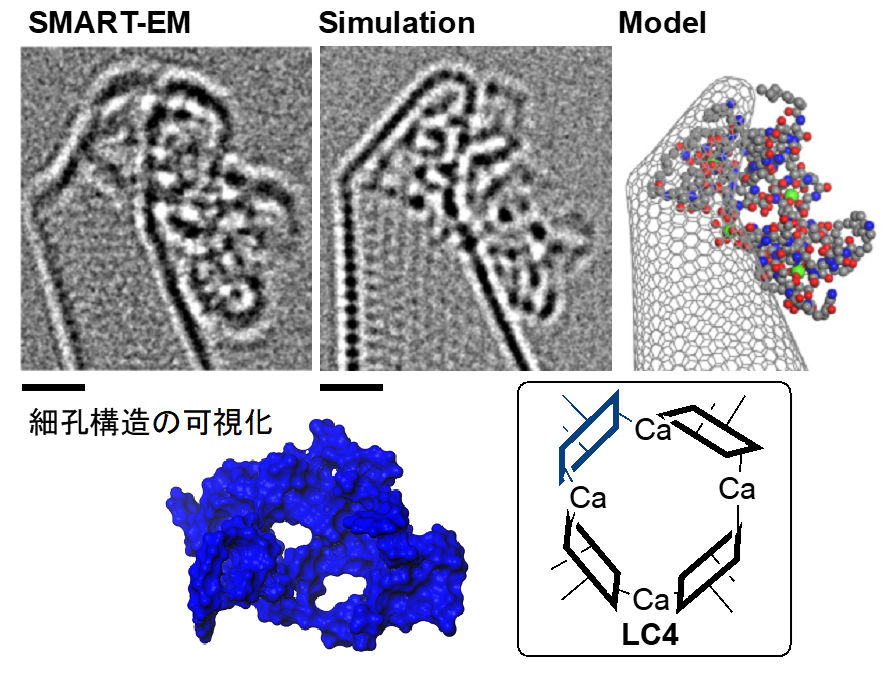
Figure 6: Structure of the DP tetramer. The blue structure is the computationally determined molecular surface and shows pores. Scale bar is 1 nanometer.
Structural analysis is fundamental to scientific research. This study not only provides a structural analysis method for complex compounds such as peptides, but also demonstrates the possibility of studying peptide aggregation behavior from the molecular level, which has been essentially unattainable. The information on the shape of molecular species, which could not be obtained by conventional methods, is expected to have a wide range of applications in the future, including drug design and material development. We expect that this method will not only provide a new tool for analyzing self-assembly phenomena, but also stimulate scientific interest in young people based on the basic fact of "looking at molecules.
The results of this research were supported by Grants-in-Aid for Scientific Research (Issue No. JP19H05459, JP21H01758, JP22K14704) and the Kanamori Foundation Research Grant. In this study, we used an atomic-resolution transmission electron microscope (JEOL JEM-ARM200F), which was introduced by the International Science Innovation Center Development Project and is a shared instrument operated by the Institute of Molecular & Life Innovation, The University of Tokyo.
Journals
-
Journal name Journal of the American Chemical Society (JACS) Title of paper Time-Resolved Atomistic Imaging and Statistical Analysis of Daptomycin Oligomers with and without Calcium Ions Author(s) Takayuki Nakamuro*, Ko Kamei, Keyi Sun, Jeffrey Bode, Koji Harano, Eiichi Nakamura*, and Eiichi Nakamura DOI Number 10.1021/jacs.2c03949
Abstract URL
Terminological Explanation
Note 1 Multidrug-resistant bacteria
Multidrug-resistant bacteria are bacteria that are resistant to two or more antimicrobial agents with different mechanisms of action. Since 2000, there has been little development of antimicrobial agents with a new mechanism of action, and thus, the understanding and suppression of multidrug-resistant bacteria has become a serious problem in the medical field. Understanding and controlling the spread of resistant bacteria is important because few antimicrobial agents with new mechanisms have been developed since 2000. ↑up
Note 2 Molecular fish hook
A synthetic chemical modification of the surface of carbon nanotubes (Note 5 ), an essential technique for selectively trapping single molecules of interest for observation. See press release "Successful Direct Structural Analysis of Trace Intermediates in Chemical Reactions" ( Aug. 23, 2019 ). ↑up
Note 3 Methicillin-resistant Staphylococcus aureus (MRSA )
MRSA is one of the representative resistant bacteria, and about half of Staphylococcus aureus detected in medical facilities are said to be MRSA. daptomycin, which came into clinical use in Japan in 2011, is used as the first-line drug for bacteremia and infective endocarditis. See "Guidelines for the Treatment of MRSA Infections, Revised Edition 2019 (Committee for the Preparation of Guidelines for the Treatment of MRSA Infections, ed. ↑↑
Note 4 High-speed high-resolution electron microscopy
A transmission electron microscope with the capability to distinguish and observe each individual atom. Transmission electron microscopes are microscopes that use electron beams with wavelengths shorter than that of light. The shape of a material can be visually determined by forming an image with the electron beam transmitted through the material. Recent advances in aberration correction technology and cameras have made it possible to take images with atomic resolution and submillisecond time resolution even with electron microscopes using low acceleration voltages, which are suitable for observing organic materials. ↑up
Note 5 Carbon nanotube (CNT)
Discovered by Professor Sumio Iijima in 1991, CNT is the fifth carbon material after diamond, amorphous, graphite, and fullerene. Graphene sheets, which consist of a single layer of carbon, have an ultrafine tube-like structure with a diameter of one nanometer (one billionth of a meter) to several nanometers rounded. Carbon nanotubes exhibit a wide variety of electrical, mechanical, and chemical properties depending on their curvature, thickness, and edge conditions, and are attracting attention as an essential nanotechnology material for next-generation industries. ↑up


