Disclaimer: machine translated by DeepL which may contain errors.
Epigenome Writing Style Unraveled by Machine Learning
Keiyo Oya (Doctoral Student (at the time of research), Department of Biological Sciences)
Tetsuhito Kakutani ( Professor, Department of Biological Sciences)
Soichi Inagaki, Associate Professor, Department of Biological Sciences

![]()
The genome, which is the blueprint of all living organisms, is described by the order of the four types of bases in DNA. The genome is like a dictionary that covers the structures of all the proteins that make up the organism. In many organisms, the dictionary of the four bases of DNA is supplemented with information called the epigenome, which looks like a sticky note or a fluorescent marker. The molecular entities of the epigenome are the methylation of DNA itself and various chemical modifications such as methylation and acetylation of proteins bound to DNA. One of the roles of the epigenome is to provide efficient access to genomic information by placing epigenomic sticky notes on necessary and unnecessary parts of the genome, since the genome is large and not all the information is necessary. The epigenome, like the genome, is sometimes inherited during cell division or when an individual bears offspring, but it can also be reattached sequentially depending on the growth environment and other circumstances. The epigenome is not only important for the development of an individual, but is also known to sequester the function of harmful genes in the genome and to be deeply involved in diseases such as cancer.
An important question in the field of epigenome research is how and where epigenomic information is written and erased. This means that sticky notes should be placed in the appropriate places. In other words, how does the system allow sticky notes to be placed in the appropriate places?
To elucidate this mechanism, we have studied epigenetic modifications, which are common to many organisms including plants and animals. Since the enzyme protein that writes the modification should approach the target region when writing, we expected that we would be able to determine the writing rule by examining the region in the genome where the modification is present. Therefore, we experimentally investigated the existence patterns of writing enzymes and searched for how the patterns were determined by using machine learning. As a result, it became clear that there are some enzymes that write the same epigenome modification but have different rules for the writing location.
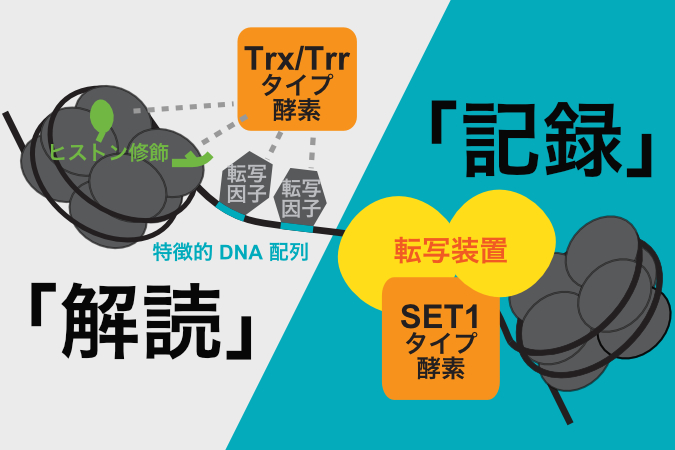
It is suggested that some enzymes write the epigenome when genetic information is read (transcribed), while other enzymes use other epigenomic information or DNA sequence information as a cue. In other words, one enzyme writes the epigenome as a "record" of gene transcription, while another enzyme writes the epigenome by "decoding" the genome and other epigenomic information (Figure). It is also clear that these two broad modes of epigenome writing are shared by evolutionarily distant organisms, plants and animals. Repeated experiments and analyses as in this study are expected to unravel the complex mechanisms of epigenome regulation.
The results of this study were published in Nature Communications 13, 4521 (2022) by S. Oya et al.
(Press release on August 11, 2022)
Published in the November 2022 issue of Faculty of Science News
Communicating to Faculty Research Students on the Frontiers of Research>