Disclaimer: machine translated by DeepL which may contain errors.
~ Message from a graduate student~.
Computational solutions to the evolutionary and developmental processes of life
 |
| Naoki Konno |
| Department of Biological Sciences, 1st Year Doctoral Student |
| Birthplace Kanagawa Prefecture |
| High School Komaba High School attached to the University of Tsukuba |
| Faculty Faculty of Science, The University of Tokyo Department of Biological Sciences |
Life is a very complex system consisting of many cells and gene functions. I love living things and have been interested in biology since junior high school and high school, but as I learned more about biology, I began to wonder how such a well-organized system could be created. The first step in answering this question is to understand how a system can be so well designed. To answer this question, first, it is necessary to comprehensively elucidate the genealogy of the processes by which life is formed, namely evolution (the process by which diverse life forms emerge from a single common ancestor) and development (the process by which a multicellular adult organism is formed from a single fertilized egg). Therefore, while studying in the Department of Information Science in the Faculty of Science at TUFS, I began research on information analysis techniques for studying the genealogy of evolution and development.
The evolutionary lineage of life can be expressed as an evolutionary phylogenetic tree, i.e., a tree structure with repeated bifurcations (speciation) (Figure 1). The genealogy of development can also be expressed as a cell lineage, a tree structure that repeats bifurcation (cell division). Both of these evolutionary and developmental genealogies can be inferred from DNA sequence information using a computer. For example, in the case of evolution, we can estimate genealogical information based on the degree of relatedness between species by examining which species and which species have similar DNA sequences. Similarly, for developmental processes, it is now possible to infer genealogy by comparing the DNA sequences of the cells that make up the adult organism.
However, the computation to estimate the genealogy takes a very long time, because the number of possible tree structures is not known. This is because the number of possible tree structures increases exponentially as the number of DNA sequences increases. Therefore, it is difficult to estimate the genealogy for more than about 1 million DNA sequences using conventional computational techniques, and this is a major barrier to elucidating the whole picture of the evolutionary and developmental processes.
To solve this problem, I developed FRACTAL, a computational technique that can rapidly estimate huge phylogenetic and cellular genealogical information, by repeating the computational cycles shown in Fig. 2 to determine the tree structure from the root to the end, while gradually bundling a large number of DNA sequences into smaller and smaller closely related groups (the computational process is called (The computational process is like a "fractal"). In this process, the computation was greatly accelerated by allowing multiple computers to process different groups of sequences simultaneously in parallel. As a result, they succeeded in accurately estimating the genealogy of 235 million sequences, more than 200 times the conventional limit, in less than two days, and reported it as the lead paper (press release on January 7, 2022, available at https://www.s.u-tokyo.ac.jp/ja/press/2022/7702/).
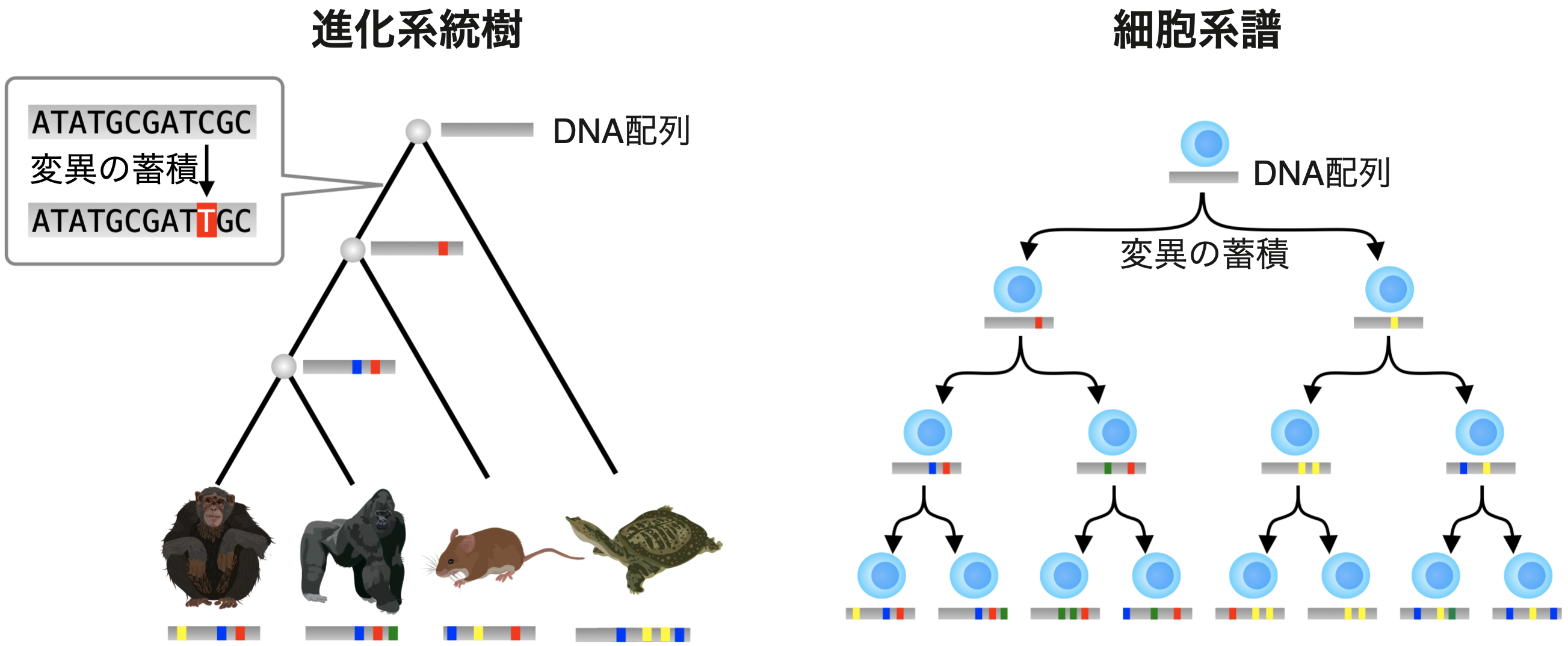
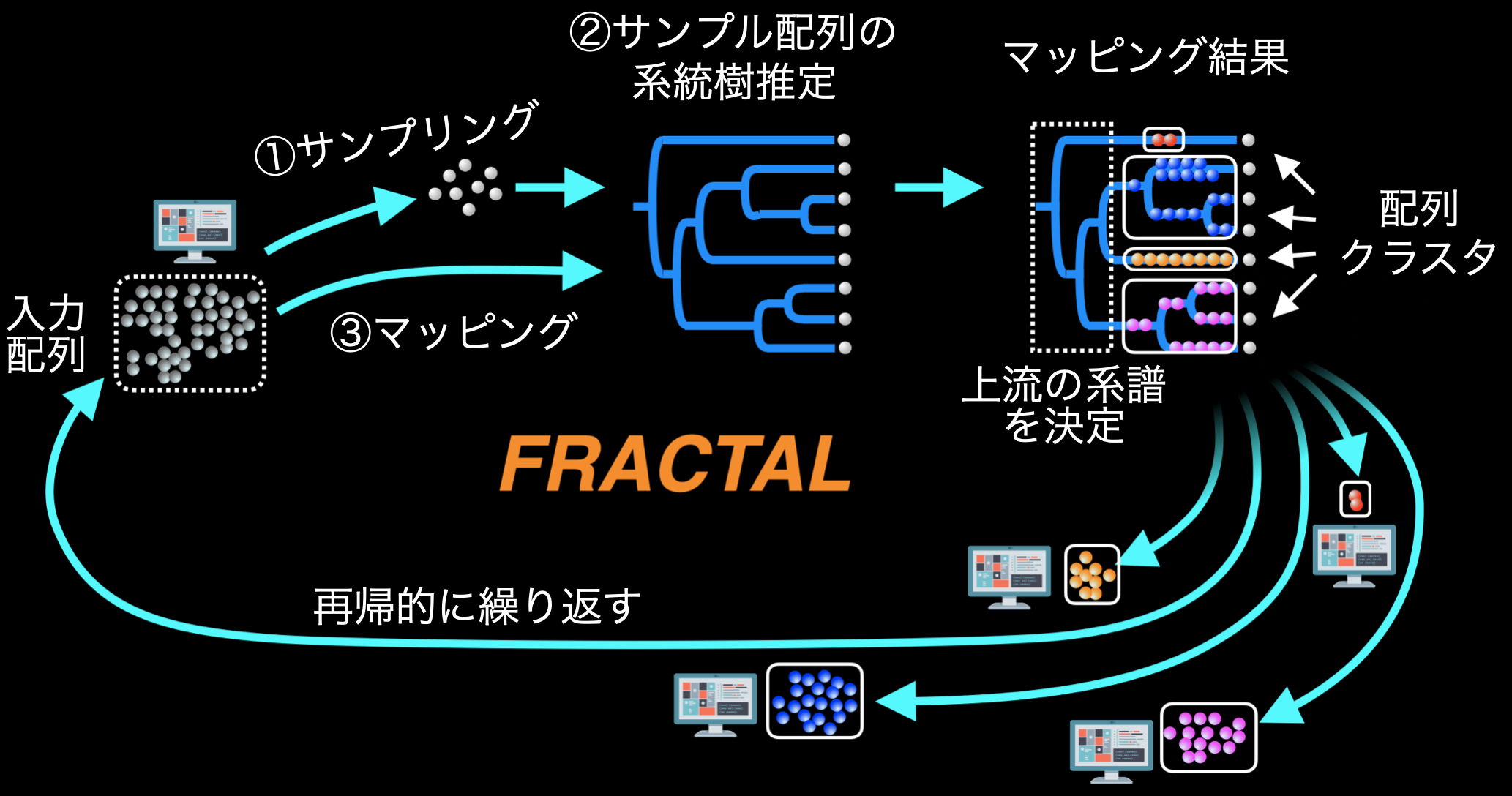
Figure 1 (top) Evolutionary phylogenetic tree and cell lineage. Illustration of organisms: © 2016DBCLS TogoTV / CC-BY-4.0
Figure 2 (bottom) FRACTAL workflow. The computational cycles shown in the figure are repeated to progressively subdivide sequences into groups that are closely related to each other, while at the same time genealogies are determined upstream
But how about this? Even if the phylogenetic tree and cellular lineage were comprehensively clarified, would that be enough to make us feel that we have understood the process of creating the system of life? I am sure that we would not. The genealogy is too complex for the human mind to comprehend. What we need next is to find some simple rule behind the phenomena we have comprehensively revealed. Therefore, I am currently conducting research to find patterns in the revealed evolutionary processes by using computational techniques called machine learning. In particular, I am focusing on the evolution of microorganisms called prokaryotes, and investigating what genes tend to be acquired or lost in what order. If the rules of evolution are understood, future life can be created. If we can understand the rules of evolution, we may be able to predict the future evolution of life (e.g., the emergence of drug-resistant bacteria). We are working daily toward the realization of this goal.


