Disclaimer: machine translated by DeepL which may contain errors.
Capturing the Evolution ofBacterial Genomes in the Laboratory
Yuki Kanai (Second-year Doctoral student, Department of Biological Sciences)Mitsuyoshi Tsuru (Project Assistant Professor, Universal Biology Institute)Chikara Furusawa (Professor, Department of Physics, Universal Biology Institute) |

![]()
Living organisms not only possess genes but also need to use them appropriately to survive. For example, in the wall cells of the stomach, several genes related to the production of stomach acid out of tens of thousands of genes in the genome need to function in accordance with the timing of the arrival of food. This "regulation of gene expression" to express the right group of genes at the right time and in the right amount is essential for all living organisms, from unicellular organisms to humans.
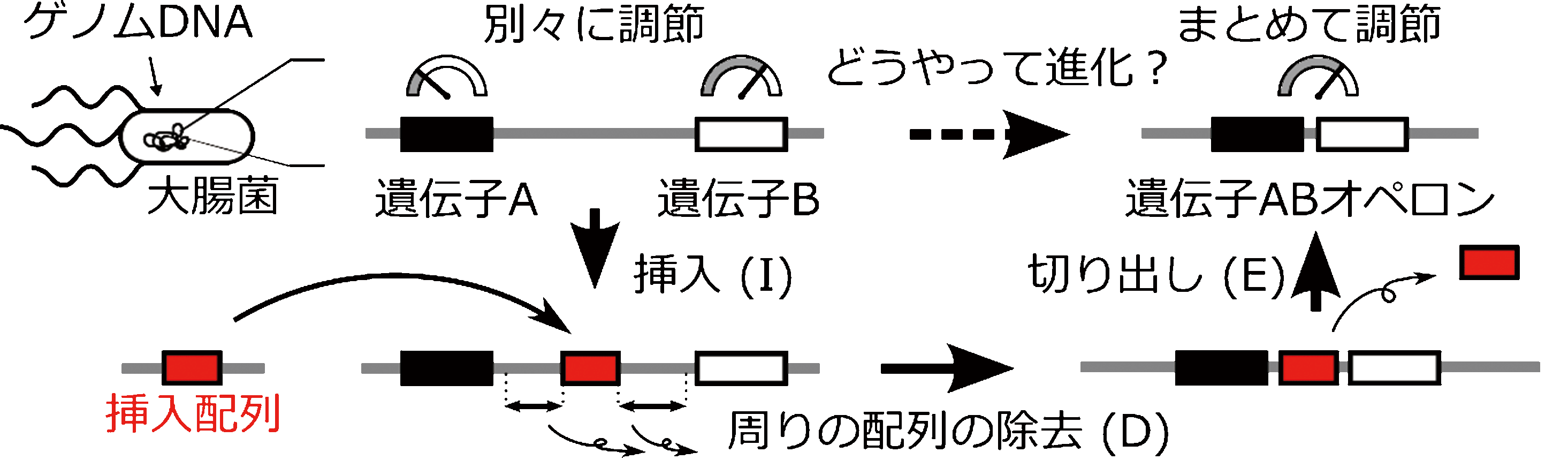
In 1961, François Jacob and Jacques Monod, through their studies of Escherichia coli, predicted that functionally relevant genes are co-located on the genomic DNA and are regulated and manipulated in a coherent manner. This unit of operation, which they called an operon, is considered one of the basic principles of gene expression regulation. However, it is not yet known how multiple genes that were originally located far from each other evolved into an operon (Figure).
 |
|
| Figure: IDE hypothesis of operon formation. Two genes that are regulated separately come closer to each other by the activity of the insertion sequence and are regulated together as an operon. | |
In this study, we proposed a new theory that may be a breakthrough. Many bacteria have insertion sequences, which are sequences (transposons) that move along the genomic DNA. Strangely enough, during the evolution of insect gut bacteria, it is known that the number of these insertion sequences increases explosively to several hundred. To understand this evolutionary process, we attempted to reproduce the explosive increase in insertion sequences using Escherichia coli, which has genetic information similar to that of the ancestor of insect gut bacteria. We prepared E. coli with insertion sequences with high self-replication activity and allowed them to evolve in the laboratory, and unexpectedly, we observed that the insertion sequences removed sequences around themselves.
Based on this phenomenon, we considered the following (Figure). In the ancestral genome, the insertion sequence may have inserted itself between genes located far apart (Insertion), removed surrounding sequences (Deletion), and finally cut itself out (Excision), resulting in the formation of an operon. To support our hypothesis that an operon could be generated, we prepared E. coli with an insertion sequence between two genes. When we incubated the E. coli overnight, a novel operon was indeed formed.
This discovery was the result of our research on the universal nature of evolution through the evolution of E. coli, and our exposure to insect gut bacteria and insertion sequences. We never know where the hints of discovery may be found.
The results of this study were published in Y. Kanai et al. Nucleic Acids Research 50, 1673 (2022).
(Press release, January 24, 2022)
Published in Faculty of Science News, May 2022
Communicating to Faculty Research Students >


