Disclaimer: machine translated by DeepL which may contain errors.
Discovering New Knowledge about Cancer by Deep Learning Feature Extraction
Tatsuhiko Tsunoda, Professor, Department of Biological Sciences |


Our research focuses on elucidating the mechanism of cancer development using molecular data from cancer patients, and on methods for identifying the individuality of cancer in each patient and selecting the optimal treatment. The molecular data referred to here include the transcriptome, which is the genome and RNA transcribed from the genome, the proteome, which refers to all proteins expressed in the cell, the metabolome (total metabolic substances contained in the body), and the epigenome, which modifies genetic information. The study that comprehensively brings all of these together is called omics. Since the individuality of cancer is determined by the complex relationships among these many omics, it is difficult to handle with conventional statistics.
Deep learning is based on a deep neural network (DNN) with multiple intermediate layers of nonlinear functions, which can handle complex patterns and is excellent for image processing, etc. I have been focusing on deep learning, but since omics is not an image but high-dimensional data, after much trial and error to find a good method, I came up with the following After much trial and error, I realized that it would be better if the omics data could be represented in two dimensions and the molecules could be re-mapped so that molecules that behave similarly among many specimens are close to each other. In this way, the omics data made into images were handled by deep learning, and the cancer type could be determined with high accuracy (DeepInsight method published in 2019). So what does DNN look for to identify cancer?
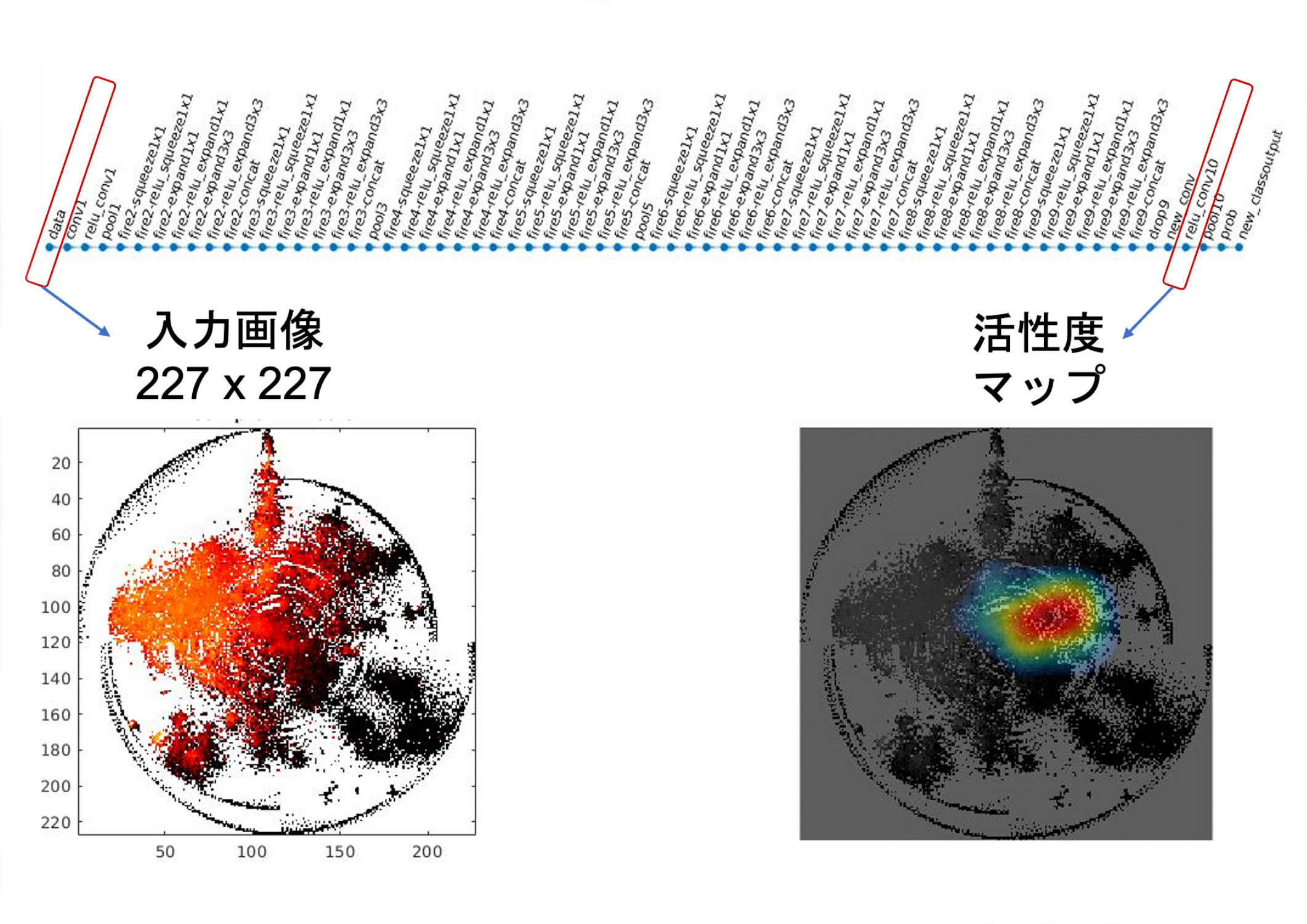
In DNNs, after training, various features are aggregated in an intermediate layer near the output layer (right side of the figure), and these features are combined to discriminate images. We considered the analysis of this layer to be the key. We used a technique called CAM (Class Activation Mapping) for the analysis. When an image is input after training (left side of the figure), the output fk (x,y) in ( x,y ) coordinates on this intermediate layer is calculated for each feature k (the "activation map" on the right side of the figure). In addition, a weight wk representing the degree to which each feature k supports the discriminant target (e.g., lung cancer) is also calculated in the DNN. Then, for each feature, fk (x,y) - w is calculated by deep learning feature extraction to discover new knowledge about cancer, and finally all the features are added together. Then, when lung cancer is judged, the degree of contribution of each pixel in the input image is known, i.e., which pixel was looked at to make the judgment. When the DeepFeature method was applied to an experiment to discriminate and predict 10 types of cancer, it was found that genes commonly involved in cancer, such as epithelial-mesenchymal transition, coagulation, angiogenesis, hypoxia, and inflammatory response, were observed. In addition, we newly found that the DeepFeature method looks at extracellular matrix structures such as collagen, receptor tyrosine kinase signaling via phosphorylation of protein tyrosine residues, G protein-coupled receptor (GPCR) ligand binding, and other signaling pathways.
Deep learning has a "black box" problem in which the path to the conclusion is not known, but with this research as a clue, it will be possible to break free from this problem and discover the process. In the future, we aim to elucidate the complex and dynamic mechanism of carcinogenesis and to determine the treatment for each patient by looking at the detailed differences in the "face" of cancer.
The results of this study were published in A. Sharma et al.
(Press release, August 19, 2021)
Published in Faculty of Science News, January 2022
Communicating to Faculty Research Students on the Frontiers of Research>