Disclaimer: machine translated by DeepL which may contain errors.
Commonality and Individuality of Neural Activity Revealed by Whole Brain Measurement
Yu Toyoshima, Associate Professor, Department of Biological Sciences
Yuichi Iino, Emeritus Professor, The University of Tokyo*Former Professor, Department of Biological Sciences
Neurons that make up the cranial nervous system are connected to each other to form neural circuits, and process information by exchanging signals of neural activity. How are activity patterns generated in actual neural circuits?
Since the activity of individual nerves is determined by signals sent from upstream nerves, if we can observe all neural activity simultaneously, we can get closer to the mechanism by which they interact with each other to produce each other's neural activity.
From this point of view, we have been working on "whole-brain measurement" and have found that neural circuits with small individual differences generate neural activity with large individual differences.
![]()
In the brain and nervous system, many neurons are connected and exchange signals with each other to process complex information. By clarifying the connections between cells and examining neural activity, we can elucidate the mechanism of information processing. However, the human brain is extremely complex, consisting of tens of billions of neurons, and it is difficult to elucidate the connections between individual cells. On the other hand, the nematode C. elegans, a small animal with a body length of approximately 1 mm, has a compact nervous system consisting of 302 neurons, and the connections between neurons are already known. Because of its small size and transparency, C. elegans can express fluorescent proteins that change brightness in response to neuronal activity, making it possible to observe neuronal activity while the animal is alive.
To take advantage of these advantages of nematodes and realize "whole-brain imaging," we have been working with Professor Ken Ishihara of Kyushu University, Professor Ryo Yoshida of the Institute of Statistical Mathematics, and Lecturer Tadashi Iwasaki of Ibaraki University to develop high-speed fluorescence microscopes, image analysis techniques, and nematode strains suitable for such imaging. They have succeeded in simultaneously measuring the activity of approximately 180 neurons in the head at the single-cell level.
In C. elegans, individual neuron types can be distinguished, so the activity of the same neuron can be compared between individuals. However, the time series of whole brain neuronal activity obtained in this study showed large differences when compared between individuals reared under the same conditions, and it was difficult to find common neuronal activity among individuals. Since individual differences in cell-cell connections in the C. elegans nervous system have been thought to be small, the large individual differences in neural activity were unexpected. In this study, we developed a new mathematical analysis method and extracted common neural activity motifs from the time series of neural activity of each individual. The types of motifs and their timing differed from individual to individual, and these differences appeared as individual differences in the time series.
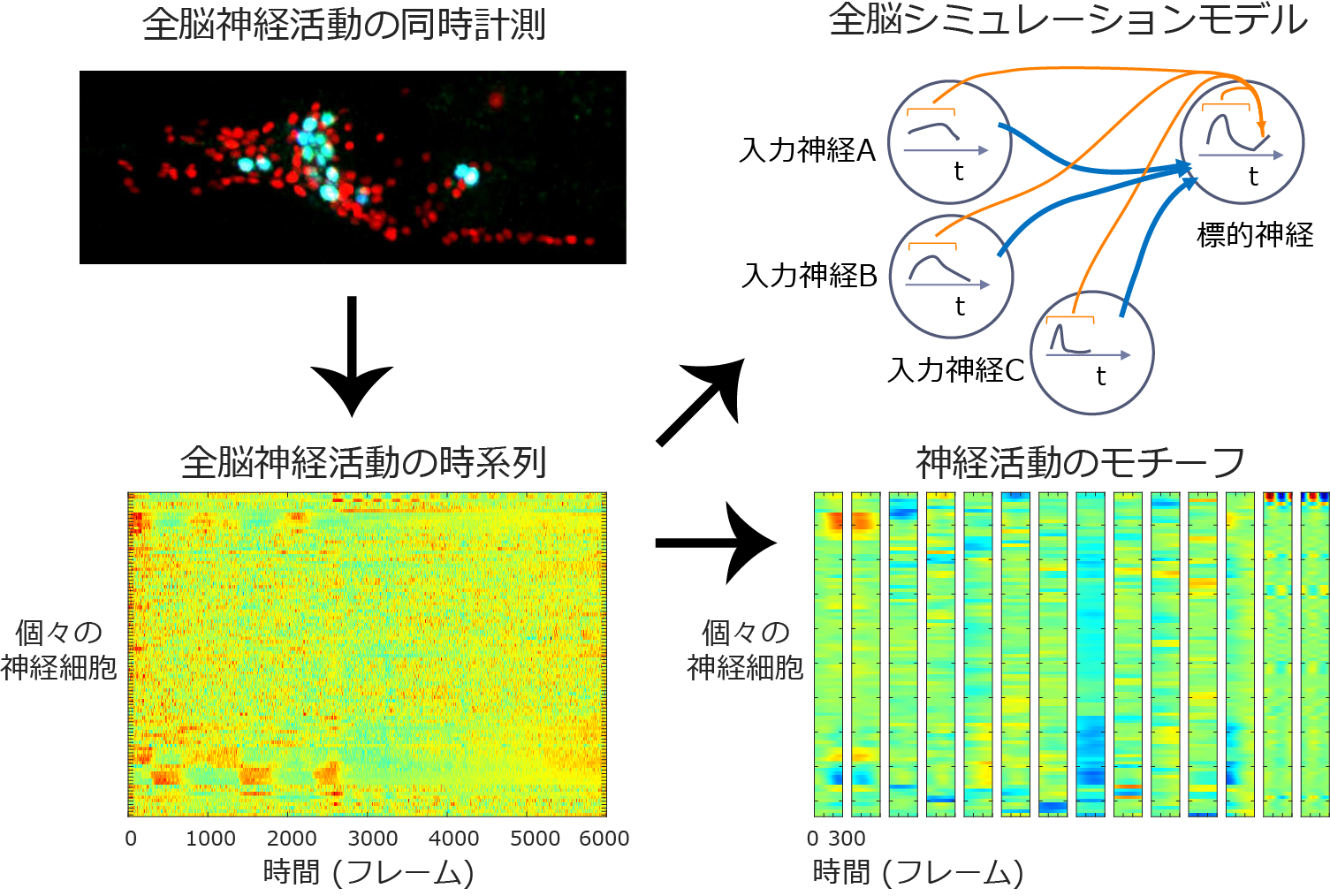 Fluorescent proteins (shown in red and light blue) were expressed in the nuclei of neurons in the head of C. elegans, and the neuronal activity was observed as three-dimensional moving images. This image was analyzed to extract whole brain neural activity. From the whole-brain neural activity, we developed a method to find motifs of neural activity common among individuals and a whole-brain simulation model.
Fluorescent proteins (shown in red and light blue) were expressed in the nuclei of neurons in the head of C. elegans, and the neuronal activity was observed as three-dimensional moving images. This image was analyzed to extract whole brain neural activity. From the whole-brain neural activity, we developed a method to find motifs of neural activity common among individuals and a whole-brain simulation model.
Since individual neurons are activated by inputs from upstream nerves, future neural activity can be predicted if the strength of the input at a given point in time is known. The strength of the input can be calculated by knowing the activity of upstream nerves and the strength of cell-to-cell connections. In C. elegans, however, the presence or absence of intercellular connections was known, but the strength of these connections was not. Therefore, we developed a time series prediction model by using the whole brain neural activity obtained in this study to estimate the strength of cell-cell connections. This model successfully reproduced the characteristics of the whole-brain neural activity obtained in the experiment, making it possible for the first time in the world to perform a whole-brain simulation based on actual whole-brain neural activity. The estimated values of the strength of intercellular connections revealed that the strength of connections between sensory nerves and downstream interneurons varied greatly from individual to individual.
Thus, this study clarified how neurons interact with each other in actual neuronal circuits. The methodologies developed and the findings obtained in this study are expected to be useful as fundamental tools for understanding how actual neuronal circuits process information from the external world.
The results of this study have been published in Y. Toyoshima et al, PLOS Computational Biology, 20, e1011848 (2024).
(Press release March 16, 2024)
Published in The Rigakubu News, July 2024
The Frontiers of Research for Undergraduates>


