DATE2023.01.10 #Press Releases
Elucidating the mechanism that sorts out abnormal ribosomal traffic jams
Disclaimer: machine translated by DeepL which may contain errors.
--Visualization of complex movements that control quality control using high-speed AFM
The University of Tokyo
Nagoya University
National Institutes of Natural Sciences, Center for the Exploration of Life
Japan Science and Technology Agency
Announcement Summary
A research group led by Associate Professor Yoshitaka Matsuo and Professor Toshifumi Inada of the Department of RNA Regulation, Institute of Medical Science, The University of Tokyo, in collaboration with Professor Takayuki Uchihashi of the Graduate School of Science, Nagoya University, who is also Visiting Professor at the National Institute of Natural Sciences, Department of Frontier Sciences, has elucidated the mechanism by which ribosomes sort out abnormal traffic jams. The research group has elucidated the mechanism that sorts out abnormal traffic congestion in ribosomes.
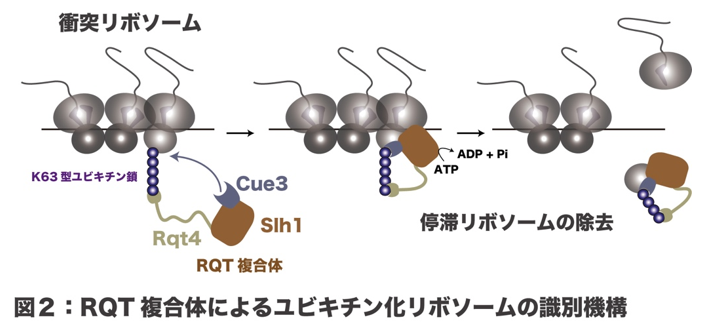
Ribosomes are devices that read genetic information from mRNA and perform "translation" to synthesize proteins, and when a ribosome stops translation, subsequent ribosomes collide with it, causing a ribosomal traffic jam. When ribosome traffic jams accumulate in the cell, various stress responses are induced. On the other hand, to prevent excessive stress responses, cells have a quality control mechanism that resolves ribosomal traffic jams. Hel2, a quality control factor, identifies ribosome collisions and adds ubiquitin as a marker for abnormal translation. This ubiquitination serves as a marker and the RQT (Ribosome Quality control Trigger) complex resolves the traffic jam by forcing the colliding ribosomes to dissociate their subunits. Ubiquitination of ribosomes is essential for the removal of collision ribosomes, but the molecular mechanisms that identify it are poorly understood.
Using biochemical methods, this research group found that K63-type ubiquitin chains are formed on collision ribosomes, which are identified by Cue3 and Rqt4, the component proteins of the RQT complex. Furthermore, using high-speed AFM, which can visualize the dynamics at the single molecule level, they succeeded in visualizing the movement of the RQT complex, especially the highly motile naturally degenerate regions.
These results are expected to lead to a better understanding of the pathogenesis of neurodegenerative and other diseases that are thought to be caused by a breakdown of the quality control mechanism and to the development of novel therapeutic strategies.
This research was supported by the Japan Science and Technology Agency (JST) through the PRESTO "Dynamic Higher-Order Structure of Cells" project (project number: JPMJPR21EE, PI: Yoshitaka Matsuo), the Grant-in-Aid for Scientific Research by the Japan Society for the Promotion of Science (project numbers: 21H00267, 21H05710, 22H02606, and H02606, Yoshitaka Matsuo; 21H01772, 21H00393, Takayuki Uchihashi; 19H05281, 21H05277, 22H00401, Toshifumi Inada), Japan Agency for Medical Research and Development (AMED-CREST project number: 20gm1110010h0002, PI: Toshifumi Inada). The results of this research are expected to be published in the January 10, 2023 issue of the Journal of Medical Research and Development.
The research results were published online in the British scientific journal Nature Communications at 7 p.m. on Tuesday, January 10, 2023 (10 a.m. UK time).
For more information, please visit the website of the Institute of Medical Science, The University of Tokyo.


