DATE2022.08.23 #Press Releases
Mechanism and significance of time-dependent transcription of the clock gene Bmal1.
Disclaimer: machine translated by DeepL which may contain errors.
Yasuko Abe (Doctoral Student, Department of Biological Sciences at the time of research)
Hikaru Yoshitane (Assistant Professor, Department of Biological Sciences at the time of the research)
Associate Professor, Department of Biological Sciences, Tokyo Metropolitan Institute of Medical Science
Yoshitaka Fukada (Professor, Department of Biological Sciences at the time of research / Emeritus Professor, University of Tokyo)
Key points of the presentation
- We have shown that feedback regulation of the clock gene Bmal1(Note 1) via its transcriptional rhythm stabilizes the biological clock through three levels of hierarchy: cultured cells, individual mice, and mathematical simulations.
- By deleting the transcriptional regulatory sequence upstream of the clock gene Bmal1 by genome editing technology, the transcription rhythm of Bmal1 to mRNA (Note 2) was successfully eliminated.
- By clarifying the molecular mechanism by which the body clock robustly maintains the 24-hour cycle rhythm, it is expected to enable the development of new methods for medical and pharmacological interventions for sleep disorders, jet lag, and other disorders.
Summary of Presentation
In the mammalian biological clock mechanism, an oscillatory model that combines two types of feedback regulation via gene transcription and translation has been proposed. The group led by Yasuko Abe (then Graduate Student), Hikaru Yoshitane (then Assistant Professor), and Yoshitaka Fukada (then Professor) at the Graduate School of Science, The University of Tokyo, focused on the clock cis-element (Note 3), which is the most important regulator of the expression rhythm of the clock gene Bmal1, and by using genome editing technology (Note 4) deleted this element This is a new approach to eliminate the second feedback loop of the biological clock. Furthermore, by combining experiments using cultured cells and individual mice and mathematical simulations, we clarified that the evolutionarily conserved second feedback loop mechanism is important for maintaining a robust biological clock cycle in a fluctuating environment. This achievement is a very important cornerstone for elucidating the molecular mechanism of the body clock, which stably ticks a rhythm of approximately 24-hour cycles.
Announcement
Many organisms on the earth have acquired an internal clock mechanism, an endogenous timekeeping system that generates a rhythmic 24-hour cycle, in order to adapt to the external environment, which oscillates with a 24-hour cycle. The body clock rhythmically regulates various physiological functions such as sleep, metabolism, and hormone secretion, and the elucidation of its molecular mechanism is an important issue. In the mammalian biological clock, an oscillatory model that combines two feedback loops via gene transcription and translation has been proposed (Figure 1).
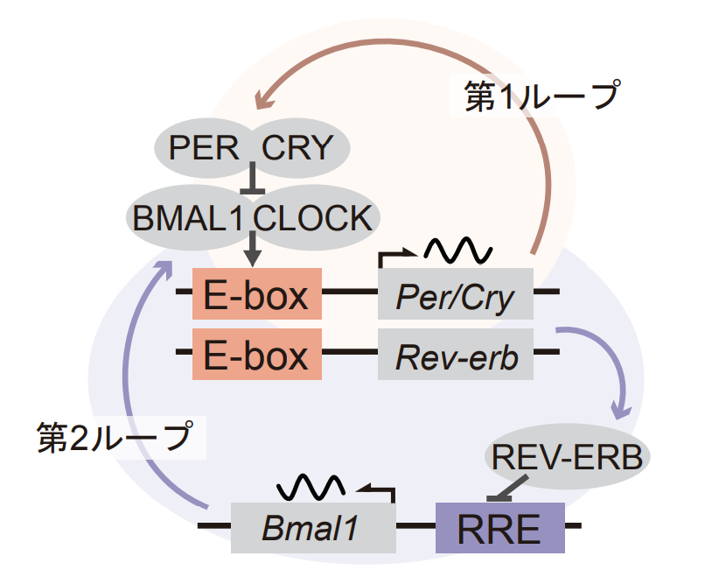
Figure 1: Conventional model of the biological clock with two feedback loops coupled. The feedback structure is established by the rhythmic transcriptional control of genes via the clock cis-elements, E-box and RRE, and the translated proteins positively or negatively regulate the activity of the clock cis-elements. The first loop is shown in orange and the second loop in blue.
In this feedback mechanism, it was known that transcription of each gene is rhythmically controlled via regulatory sequences (called clock cis-elements) upstream of the gene, but the importance of the feedback loop via each gene expression rhythm was not known.
A research group led by then Graduate Student Yasuko Abe, then Assistant Professor Hikaru Yoshitane, and then Professor Yoshitaka Fukada at the Graduate School of Science, The University of Tokyo, focused on the clock cis-element, the most important action point that controls the gene expression rhythm, and succeeded in eliminating the second feedback loop of the biological clock by using genome editing technology to delete this element They succeeded in eliminating the second feedback loop of the biological clock by using genome-editing technology. Specifically, by deleting the clock cis-element RRE, which is located upstream of the clock gene Bmal1, they generated a mutant cell line and mutant mouse individuals in which the expression rhythm of the Bmal1 gene was abolished without losing the main body of the gene (Figure 2).
Examination of the ring-rotating behavioral rhythm of the mutant mice revealed an autonomous activity rhythm even under constant-dark conditions with no light information, and no significant differences were observed between the mutant and wild-type mice. In addition, autonomous oscillations at the genetic level were continuously observed in organs and mutant cell lines of the mutant mice, indicating that the internal clock oscillations continue to be stable even in the absence of the second feedback loop via rhythmic transcription of Bmal1, a result that defies the conventional model. To explore the mechanism in more detail, we simulated the biological clock feedback regulation in the mutant using the clock mathematical model (Note 5). The results of the experiment and the mathematical model agreed well, showing that the biological clock oscillation is maintained even when the Bmal1 transcription level is constant. The simulations predicted that the phosphorylation rhythm of the BMAL1 protein (i.e., the rhythm of BMAL1 function) would be maintained despite the loss of the Bmal1 gene transcription rhythm, indicating that post-translational regulation may continue the functional rhythm of BMAL1.
Interestingly, both the mathematical model and the cell lines revealed that the amplitude of the biological clock oscillation is more likely to be weakened and the cycle more likely to be prolonged in the mutant than in the wild type when the biological clock is externally perturbed. This result suggests that a second feedback regulatory mechanism via the transcriptional rhythm of Bmal1 functions to stabilize the biological clock oscillation. We conclude that the second feedback regulation via the clock cis-element RRE upstream of the Bmal1 gene plays an extremely important role in maintaining the robustness of the biological clock's timekeeping system, which oscillates with an approximately 24-hour cycle.
The results of this study are important basic research that will help to elucidate the deeper molecular mechanisms of the body clock, which stably ticks a rhythm of approximately 24-hour cycles, and to treat various rhythm disorders, including sleep. Furthermore, the method of this research, which focuses on the clock cis-element, the regulatory sequence of the gene, rather than modifying the gene itself, is expected to have a strong impact on a wide range of research fields.
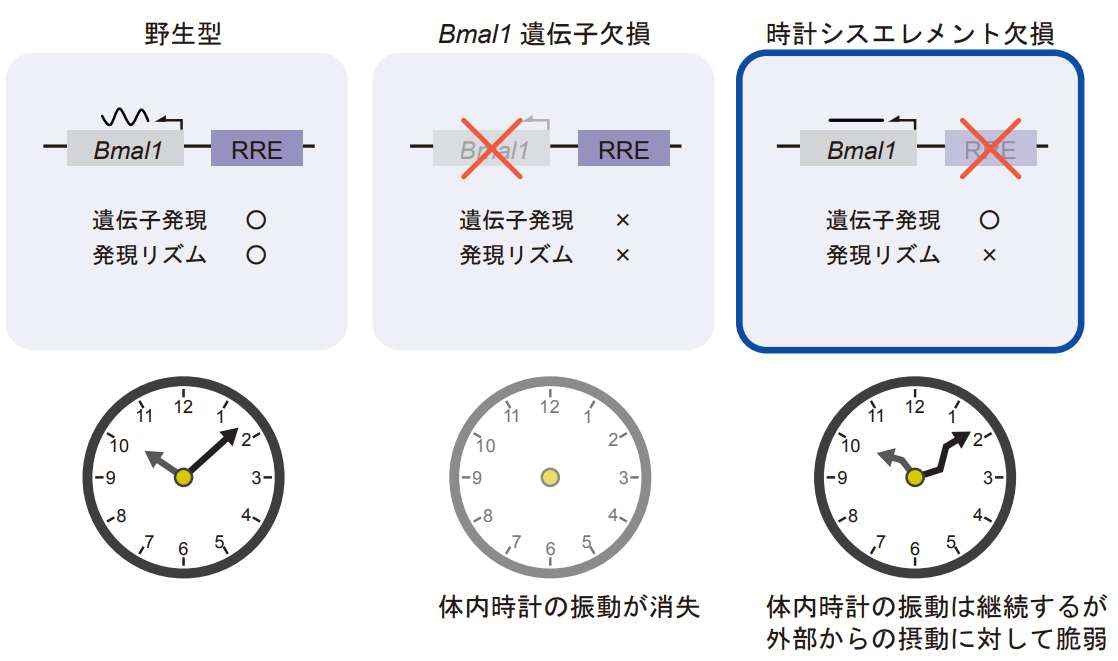
Figure 2: Relationship between rhythmic regulation of the clock gene Bmal1 gene and body clock oscillation. In the wild type (left), the Bmal1 gene is rhythmically transcriptionally regulated by the clock cis-element RRE located upstream of the genome, and stable body clock oscillations are observed; when the Bmal1 gene is deleted (center), Bmal1 is not expressed and the body clock oscillations are lost However, the significance of the rhythmic transcriptional regulation of the Bmal1 gene was not known. In this study, by deleting the clock cis-element RRE upstream of Bmal1, we succeeded in creating a state in which the Bmal1 gene is expressed but the expression rhythm is lost (right). In this case, an internal clock oscillation was observed, but it was found to be more fragile than in the wild type.
This research was conducted in collaboration with Associate Professor Jae Kyoung Kim at KAIST, Korea Advanced Institute of Science and Technology, and Professor Atsushi Feiba at Graduate School of Medicine, The University of Tokyo. Research was also conducted at the Tokyo Metropolitan Institute of Medical Science.
Journals
-
Journal name Nature Communications Title of paper Rhythmic transcription of Bmal1 stabilizes the circadian timekeeping system in mammals Author(s) Yasuko O. Abe, Hikari Yoshitane*, Dae Wook Kim, Satoshi Kawakami, Michinori Koebis, Kazuki Nakao, Atsu Aiba, Jae Kyoung Kim* and Yoshitaka Fukada* (in Japanese) DOI Number
Terminology
1 Clock gene Bmal1
A transcription factor essential for the autonomous oscillation of the biological clock and widely conserved in mammals, BMAL1 protein binds to the clock cis-element E-box together with CLOCK protein and activates transcription of neighboring genes. Cryptochrome genes, when transcribed and translated, bind to the BMAL1-CLOCK complex and repress its transcription-promoting activity. Furthermore, the Rev-erb gene, which is transcriptionally regulated rhysically by E-box, rhysically regulates transcription of the Bmal1 gene via the clock cis-element RRE. Thus, the internal clock mechanism is structured as a coupling of two transcriptional feedbacks (Fig. 1). ↑up
Note 2 mRNA
Messenger RNA, a single-stranded ribonucleic acid produced by reading (transcribing) genetic information from DNA, plays a central role in the process of protein synthesis (translation) from DNA in cells. In other words, it is located in the middle of the genetic code reading process of DNA → mRNA → protein. ↑up
Note 3 Clock cis-element
Clock cis-elements are specific regulatory sequences located upstream of genes or in intronic regions, which are activated or repressed by the rhythmic binding of transcription factors, resulting in the rhythmic 24-hour cycle of gene transcription. Known clock cis-elements include E-box, RRE, and D-box. ↑up
Note 4: Genome editing
A technology to artificially modify the genomic DNA of an organism by recognizing specific base sequences with high precision. In this study, the clock cis-element upstream of Bmal1 was deleted using the CRISPR/Cas9 system. ↑up
Note 5 Mathematical clock model
A model that mathematically reproduces the oscillation of the body clock. The regulation of gene transcription, translation, and post-translational modification through the clock cis-element is expressed in mathematical equations using many variables and parameters, enabling the simulation of the biological clock. ↑up


