DATE2021.12.10 #Press Releases
Single molecule visualization of RNA helicase Vasa at work
Disclaimer: machine translated by DeepL which may contain errors.
Yoshimi KINOSHITA (Project Researcher, Department of Biological Sciences at the time of research / currently Research Fellow, Nagoya University)
Soutarou Uemura, Professor, Department of Biological Sciences
Key Points of the Presentation
- We have visualized the working of RNA helicase (Note 1) and Vasa (Note 2) in a single molecule, and clarified that Vasa functions by oligomerization (Note 3) when it peels off cleaved RNA, and that the process is heterogeneous.
- This is the first time that Vasa has been visualized working at the single-molecule level, and this study clarifies one aspect of the mechanism of helicase activity.
- The clarification of the details of Vasa's function is expected to advance the elucidation of the function of RNA helicases in general and to realize the control of RNA silencing (Note 4).
Summary of Publication
RNA helicases are enzymes that use adenosine triphosphate (ATP) (Note 5) hydrolysis energy to pull apart double-stranded RNA and are primarily involved in RNA metabolism, transcription, translation, and mRNA splicing (Note 6). It is known that the N- and C-termini of certain helicases contain regions of high fluctuation, but it has not been clarified how these regions contribute to RNA pulling off and dissociation.
Project Researcher Yoshimi Kinoshita (at the time of the research), Professor Sotaro Kamimura, and their colleagues at the Graduate School of Science, The University of Tokyo, have succeeded in visualizing the dissociation of transposon RNA (Note 7) cleaved from piRISC, a piRNA silencing complex from silkworm, by Vasa using single molecule fluorescence imaging (Note 8) The method has been used to directly investigate the involvement of Vasa in the piRNA amplification process known as the ping-pong pathway.
As a result, we found that oligomerization of Vasa is required and that Vasa promotes the dissociation of cleaved RNA in the heterogeneous pathway. Further functional elucidation will reveal the mechanism of RNA silencing, which will allow us to control RNA expression in the future.
Publication details
RNA helicases are involved in all processes of RNA metabolism, including transcription, translation, and mRNA splicing. These RNA helicases are enzymes that use the hydrolytic energy of ATP to pull apart double-stranded RNA. On the other hand, the N-terminus of certain RNA helicases has a large fluctuating region for electrostatic interaction with the same RNA helicase, and it was thought that the interaction of this region leads to the formation of droplets in the cytoplasm. These indicate that the N-terminal portion of the region causes oligomerization of the RNA helicase, but the actual contribution of this region to the helicase activity was not known.
In this study, we focused on the function of the N-terminal region of Vasa from silkworm, an RNA helicase, which functions to pull off cleaved transposon RNA from the piRNA silencing complex (piRISC) during piRNA metabolism. This cleavage reaction is carried out by the piRNA (PIWI-interacting RNA), a noncoding RNA generated specifically in germ cells, and the PIWI (P-element induced wimpy testis) of the Argonaute subfamily (AGO and PIWI proteins). This is caused by the formation of piRISC by one of the proteins, Siwi (Silkworm Piwi). In addition, the germline has a unique piRNA amplification mechanism called the ping-pong pathway to efficiently cleave transposon RNA.
The dynamic changes that cause Vasa to dissociate cleaved RNAs via piRISC directly affect the ping-pong pathway, but the mechanism by which Vasa interacts with piRISC to cause the dissociation of cleaved RNAs was not known.
In this study, we used single molecule fluorescence imaging to visualize the interaction of Vasa with piRISC in real time and evaluate how Vasa oligomerization is involved in the ping-pong pathway. First, we labeled Vasa and transposon RNA with fluorescent dyes called TMR (green) and ATTO647N (red), respectively, and measured their behavior simultaneously (Figure 1).
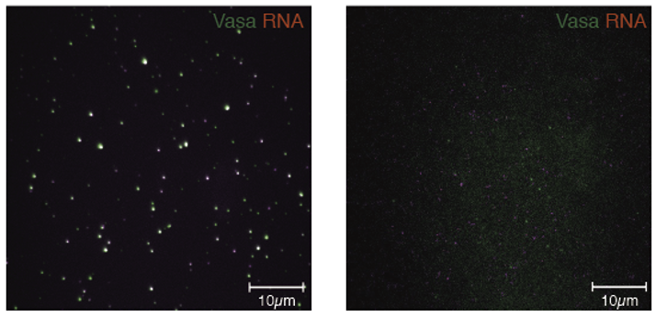
Figure 1: In the presence of Siwi, RNA (red) co-localizes with oligomerized Vasa (green) (left panel), while Vasa lacking the N-terminal region does not localize with RNA (right panel).
The results show that Vasa co-localizes with RNA in the presence of Siwi-piRISC by oligomerization in the N-terminal region, the first successful construction of a single molecule system in which Vasa forms a complex with RNA via Siwi-piRISC.
Next, we investigated how the co-localized Vasa and RNA dissociate over time. Three different dissociation patterns were observed: Vasa and RNA dissociated at the same time, Vasa partially dissociated at a different time from RNA dissociation, and Vasa partially dissociated without RNA dissociation (Figure 2).
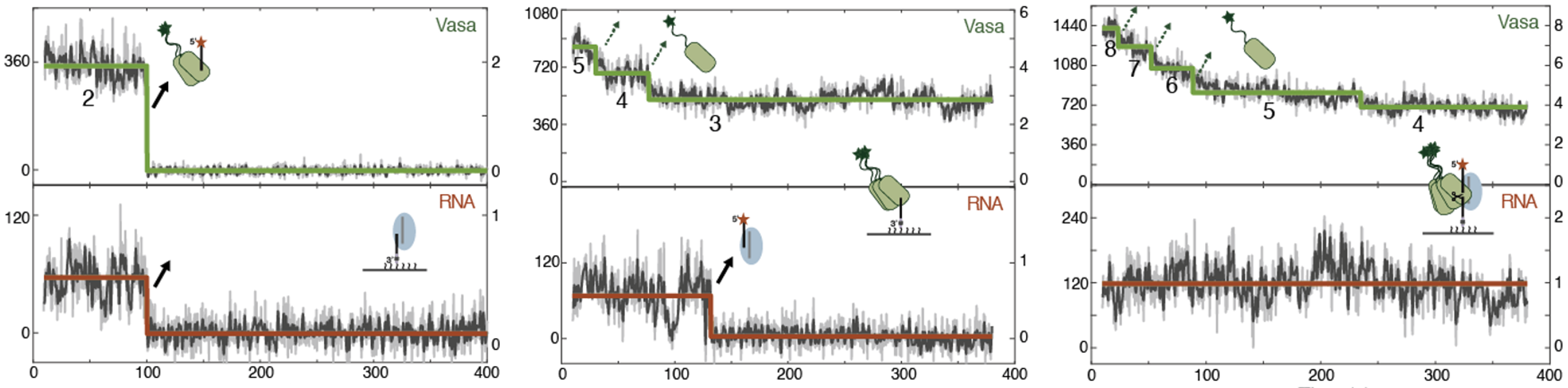
Figure 2: Time variation of the three patterns of fluorescence-labeled Vasa (green) and fluorescence-labeled RNA (red) observed simultaneously in a single molecule. (Left) Patterns in which Vasa and RNA dissociated at the same time. (Middle) Pattern of partial dissociation of Vasa at a different timing from that of RNA. (Right) Pattern of partial dissociation of Vasa without dissociation of RNA.
These experimental results led us to devise a new model in which Vasa is involved in the ping-pong pathway (Figure 3) and revealed that Vasa causes dissociation of cleaved RNA in a heterogeneous pathway, and furthermore, oligomerization of Vasa is necessary for these processes.
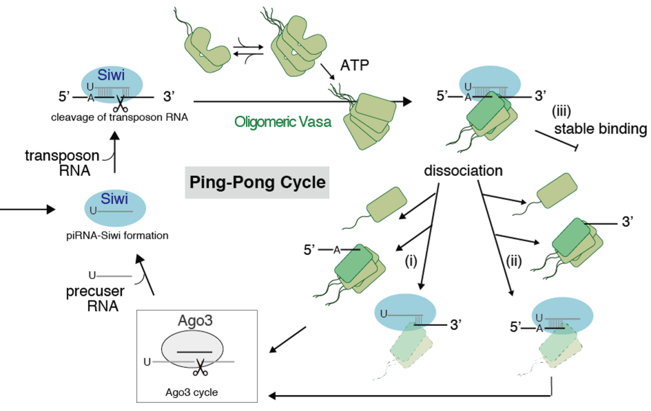
Figure 3: Ping-Pong cycle with new heterogeneous pathways. After the binding of the green Vasa oligomer to the blue Siwi-piRISC in the upper left, the following three dissociation processes proceed. (i) Oligomeric Vasa dissociates with cleaved RNA at the 5' end (ii) Oligomeric Vasa dissociates with cleaved RNA at the 3' end (iii) Oligomeric Vasa binds in a stable state. The cleaved RNA then migrates to Ago3, and Siwi-piRISC is formed again.
The results of this study suggest that coordination and heterogeneity in the timing of dissociation of oligomerized Vasa balances the dissociation in the overall ping-pong pathway. This study has provided a partial understanding of the mechanism of RNA silencing at the molecular level. Further functional analysis at the molecular level is expected to lead to future applications in the fields of medicine and drug discovery.
This research was supported by the Japan Science and Technology Agency (JST), CREST (PI: Sotaro Kamimura, PI: Mikiko Shiomi), and others.
Published Journals
-
Journal name Communications BiologyTitle of paper Heterogeneous dissociation process of truncated RNAs by oligomerized Vasa helicaseAuthor(s) Yoshimi Kinoshita, Ryo Murakami, Nao Muto, Shintaroh Kubo, Ryo Iizuka and Sotaro Uemura* (in Japanese)DOI Number 10.1038/s42003-021-02918-0Abstract URL
Terminology
Note 1 RNA helicase
A general term for proteins that have the activity to unwind the secondary structure of RNA or RNA duplex. ↑ upNote 2 Vasa
An RNA helicase that is expressed specifically in germ cells. It is thought to be involved in the common germ cell differentiation mechanism from insects to humans and in silencing of transposon RNA. ↑up
Note 3 Oligomerization
Formation of a polymer by the combination of a relatively small number (about a few) of monomers. In contrast to oligomer, polymer is a state in which a very large number (more than several hundred) of monomers are bound together. ↑up
Note 4 RNA silencing
A phenomenon in which the expression of a gene having a complementary sequence region is suppressed by a small RNA (small RNA) of 20 to 30 nucleotides in length. ↑up
Note 5 Adenosine triphosphate (ATP )
A nucleotide with three molecules of phosphate attached to the ribose of adenosine and two high-energy phosphate bonds. ↑up
Note 6 mRNA splicing
In protein biosynthesis, the process in which introns are removed and exons are joined from primary transcripts synthesized by transcription. ↑up
Note 7 Transposon RNA
A generic term for RNA transcribed from DNA of foreign origin that has the ability to freely move throughout the genome. Expression of transposon RNA can disrupt important cellular functions. ↑up
Note 8 Single molecule fluorescence imaging method
A technique to track biomolecules labeled with fluorescent molecules on a molecule-by-molecule basis. It is useful for understanding the movement, structural changes, and functions of biomolecules. ↑↑


