DATE2022.08.11 #Press Releases
Methylation for
Disclaimer: machine translated by DeepL which may contain errors.
Keiyo Oya (Department of Biological Sciences, Doctoral Program (at the time of research))
Tetsuhito Kakutani, Professor, Department of Biological Sciences
Soichi Inagaki, Associate Professor, Department of Biological Sciences
Key points of the presentation
- We found that there are two types of methyltransferases responsible for H3K4 methylation, a type of histone modification, one of which works with genome transcription and the other of which targets specific chromatin modifications and DNA sequences.
- A new research approach provided an explanation for the mechanism that introduces H3K4 methylation at the appropriate genomic location, which has been the subject of much debate.
- This achievement advances our understanding of the regulatory mechanism of H3K4 methylation, which is involved in the expression of many genes, and is expected to be applied to expression engineering in the future.
Summary of Presentations
Methylation of the fourth lysine of histone H3 protein (H3K4 methylation), a chromatin modification (Note 1 ), is highly evolutionarily conserved and distributed in the genome, especially in the highly expressed regions of the genome. There have been a number of hypotheses and discussions regarding whether H3K4 methylation promotes gene expression or is introduced as a result of gene expression, and the mechanism by which H3K4 methylation is introduced into specific genomic regions.
The University of Tokyo Graduate School of Science's Keiyo Oya, Professor Tetsuhito Kakutani, Associate Professor Soichi Inagaki and their colleagues experimentally determined the genomic distribution of each of the multiple H3K4 methyltransferases and modeled their localization patterns by machine learning to identify a type of methyltransferase that works with the gene transcription machinery (Note 2) and a type that works with a specific clock. 2 methyltransferases and methyltransferases that target specific chromatin modifications or DNA sequences to regulate H3K4 methylation.
H3K4 methylation is a modification that regulates the expression of many genes, and this study, which shows the regulatory rules of H3K4 methylation, is expected to have future applications in expression engineering and other fields.
Announcement
Chromatin modifications are essential for proper regulation of genetic information, and H3K4 methylation is a type of chromatin modification common to all eukaryotes studied, with H3K4 methylation being more densely distributed in regions of the genome that encode genes with particularly high expression levels.
There have been divergent views as to what rules are used to introduce H3K4 methylation into a particular genomic region. For example, the H3K4 methyltransferase is known to physically interact with the transcription machinery, supporting the view that H3K4 methylation is introduced as a consequence of transcription. On the other hand, in some experiments, H3K4 methylation was also observed to be introduced without prior transcription.
The team first observed the genomic distribution of H3K4 methyltransferases in the model organism Arabidopsis (Note 3 ) by means of a ChIP-seq method targeting H3K4 methyltransferases (Note 4). Next, we used machine learning to search for models that explain the localization of methyltransferases to specific genomic regions in order to determine what rules determine the localization of methyltransferases to these regions. Specifically, we trained two machine learning algorithms (random forest and support vector machine) were trained to map the localization of methyltransferases to these features.
We applied this approach to three H3K4 methyltransferases and found that one of them (SET1 type enzyme: (Note 6)) localized to a transcription apparatus binding site, suggesting that it methylates upon transcription (Figure 1 left). In contrast, where the other two methyltransferases (Trx/Trr type enzymes: (Note 6 )) localized, we found a distinctive set of chromatin modifications and a distinctive set of DNA sequences that differ from other genomic regions (Figure 1 middle).
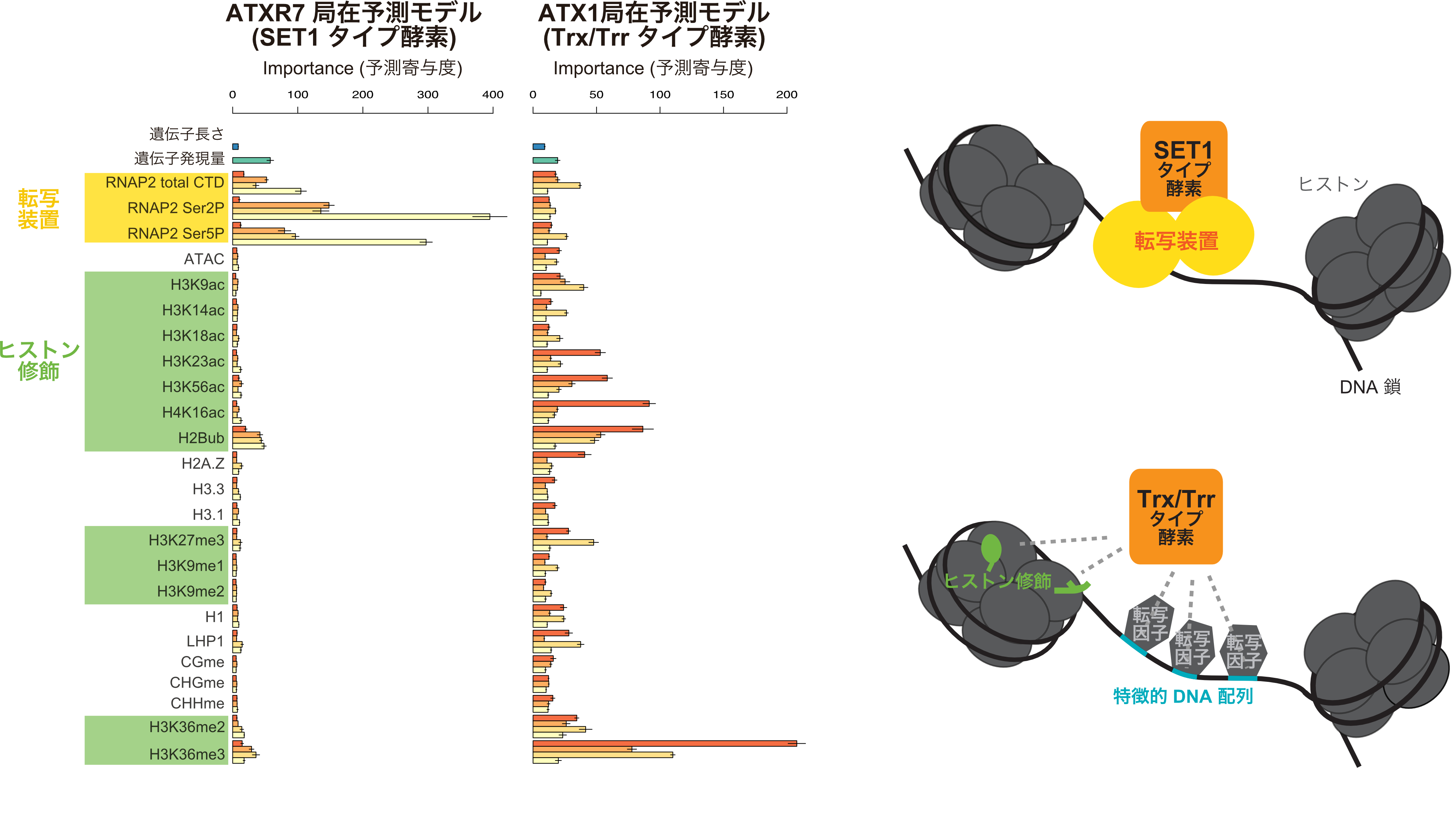
Figure 1: (left,middle) Localization of three Arabidopsis H3K4 methyltransferases, ATXR7, ATX1, and ATX2, were modeled by random forest using the chromatin feature sets shown on the vertical axis as features (cues). The horizontal axis (Importance; relative contribution) is a relative value reflecting how much each feature explains ATX(R) localization in the model. ATXR7, shown on the left, is well explained by RNAP2 (RNA Polymerase 2) transcribing genes, while ATX1 (and ATX2; figure omitted) in the middle figure is well explained by chromatin modifications. Another analysis revealed that the DNA sequences in the localized regions of ATX1 and ATX2 are also distinctive.
(Right) The model used in this study, in which H3K4 methyltransferases are evolutionarily divided into SET1 and Trx/Trr types, the former working in concert with the transcriptional apparatus and the latter with local chromatin modifications and DNA sequences as landmarks.
These results suggest that H3K4 methyltransferases have at least two modes of function. One behaves as if it "records" transcription, while the other methylates by "decoding," so to speak, specific DNA sequences and chromatin modifications. In support of these two patterns of regulation, mutants lacking the former methyltransferase showed a weaker correlation in which H3K4 methylation was distributed in regions of higher transcription, while the correlation was stronger when the latter enzyme was deficient.
To confirm the universality of this finding, we performed a similar analysis of the localization of H3K4 methyltransferase in mice using publicly available data. We found that, as in Arabidopsis, the localization pattern of SET1-type methyltransferases can be explained by transcription apparatus localization, and that of Trx/Trr-type methyltransferases by local chromatin modification. The "recording" and "decoding" mechanisms of H3K4 methylation are conserved between evolutionarily distant species, mouse and Arabidopsis (Fig. 1, right), suggesting that the two types of H3K4 methylation are conserved between evolutionarily distant species.
H3K4 methylation is a multifaceted modification that regulates transcription, and H3K4 methylation itself is dynamically regulated during development and environmental responses. In addition, diseases caused by dysregulation of H3K4 methylation are known. The mechanism for introducing H3K4 methylation demonstrated in this study has the potential for future application in technology for artificially creating desirable gene expression.
This research has been supported by JST PRESTO (JST PRESTO, Project number: JPMJPR17Q1, PI: Soichi Inagaki), CREST (Project number: JPMJCR15O1, PI: Tetsuhito Kakutani), JSPS Grants-in-Aid for Scientific Research (Project numbers: JP26221105, JP15H05963, JP 19H00995, Tetsuhito Kakutani; JP20H05913, JP22H02299, Soichi Inagaki), and JSPS Research Fellowship (JP19J21882, Keiyo Oya).
Journals
-
Journal name Nature Communications Title of paper Transcription-coupled and epigenome-encoded mechanisms direct H3K4 methylation Author(s) Satoyo Oya*, Mayumi Takahashi, Kazuya Takashima, Tetsuji Kakutani* & Soichi Inagaki* (co-authors) DOI Number
Terminology
1 Chromatin modification
DNA, the substance of genetic information, is a long, string-like molecule, but in the nucleus of eukaryotes, DNA is regularly and tightly bound to various proteins like beads. This "DNA + DNA binding protein" complex is called chromatin. The functions of chromatin include the efficient storage of long DNA in the small nucleus and the protection of DNA.
A group of chemical modifications added to chromatin is collectively called chromatin modifications. Histone proteins, the most abundant of the DNA-binding proteins, also undergo a variety of modifications at various locations on the protein, including methylation, acetylation, and ubiquitination, which are called histone modifications. Such chromatin modifications are known to be essential for proper readout, inheritance, etc. of genetic information. ↑upNote 2 Transcription machinery
A device that transcribes genetic information into RNA, a protein giant complex containing RNA polymerase that synthesizes RNA from a DNA template. ↑up
Note 3 Arabidopsis thaliana
Typical experimental plant. Arabidopsis is suitable for the study of chromatin modification regulation because of its small genome size, short generation time, easy genetic recombination, etc., and possesses most of the chromatin modifications present in eukaryotes. ↑up
Note 4 ChIP-seq
Abbreviation for Chromatin Immunoprecipitation sequencing. A method to identify the genomic location where a protein of interest binds. DNA binding to the protein of interest can be obtained by mixing an antibody that specifically binds to the protein of interest with chromatin and then collecting the DNA. The sequence of the recovered DNA is sequenced and compared with the genome sequence to determine the binding region. ↑up
Note 5: Loosening of chrom atin
It is known that the higher-order structure of chromatin plays an important role in the proper reading of genetic information. There are several methods to examine loose chromatin, such as Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq), and we used ATAC-seq data in this study. ↑up
Note 6 SET1 type enzyme and Trx/Trr type enzyme
The common ancestor of eukaryotes is thought to have had at least two H3K4 methyltransferases. The two groups of methyltransferases that originated from these two sources are referred to here as SET1-type and Trx/Trr-type enzymes, respectively. During the evolution of animals and plants, the SET1 and Trx/Trr types have undergone changes, such as changes in "forms" unique to animals and plants, duplication, and loss in some species, but the overall characteristics of the SET1 and Trx/Trr types are commonly preserved in modern animals and plants. Humans have four Trx/Trr type methyltransferase genes, and abnormalities in those genes, called MLL1 through MLL4, are one of the causes of acute leukemia. ↑


