DATE2021.08.19 #Press Releases
What does artificial intelligence stare at in genomics?
Disclaimer: machine translated by DeepL which may contain errors.
〜 ˜ How to extract features from non-image data such as genes using deep learning
Alok Sharma (Research Scientist, RIKEN Center for Biomedical Research)
Artem Ruysenko (Research Scientist, RIKEN Center for Biomedical Research)
Keith Boroevich, Technical Staff I, RIKEN Center for Biomedical Research
Edwin Vance (Graduate Student, University of the South Pacific, Engineering)
Tatsuhiko Tsunoda, Professor, Department of Biological Sciences / Team Leader, RIKEN Center for Biomedical Research
Key points of the presentation
- We developed a DeepFeature method that converts non-image omics (Note 1)datasuch as genomic (Note 2) data into image format and treats them with deep learning, and then uses deep learning to discover what genes and other features to focus on. It has shown excellent performance in experiments to predict cancer types, and at the same time, discovered novel signal pathways to discriminate cancer types.
- This is the first method in the world that enables biomedical interpretation of what deep learning focuses on when non-image data such as genomes are imaged and trained, and extracts important genes and other features.
- This research, along with prioritization of diagnostic markers, interpretation of complex multi-omics (Note 3) data, and causal discovery, could be used as a thread to promote deep learning to break out of the "black box" model and discover how conclusions were reached.
Presentation Summary
Artificial intelligence, especially deep learning (Note 4), is used in many fields and is expected to be used in genomics (Note 5 ) because it can discriminate complex patterns that cannot be captured by classical statistical methods. However, how to handle such ultra-high-dimensional, non-image data with deep learning and how to interpret the resulting deep learning models from a biomedical perspective have been completely unresolved issues. Professor Tatsuhiko Tsunoda of the Graduate School of Science at The University of Tokyo (Team Leader, RIKEN Center for Biomedical Research) and his colleagues have developed the DeepFeature method, which converts non-image data such as genomic and omics data into image format and handles them with deep learning, and also uses deep learning to discover which genes and other features to focus on. The method compresses data into an image format and handles it with deep learning. The DeepFeature method is applied to an experiment to predict cancer types, and the results showed excellent prediction performance as well as good discrimination of cancer types. The DeepFeature method was shown to have excellent predictive performance as well as the ability to discover novel signaling pathways that discriminate between cancer types. This is the first method in the world that enables biomedical interpretation of what is important in deep learning when genome and other data are imaged and trained, and extracts important genes and other features.
This research, along with prioritization of diagnostic markers, interpretation of complex multi-omics data and causal discovery, could be used as a thread to promote deep learning to break out of the so-called "black box" model and discover how conclusions were reached.
Presentation Details
Background of the research and problems in previous studies
Artificial intelligence, especially deep learning, has been used in many fields because it can discriminate complex patterns that cannot be captured by classical statistical methods, and is now expected to be used in the field of omics, which analyzes genomes and other information. In the world's first attempt to use deep learning to handle such ultra-high-dimensional, non-image data, Professor Tatsuhiko Tsunoda of The University of Tokyo's Graduate School of Science and his colleagues have previously established a new approach to convert such data into image format so that they can be learned by convolutional neural networks (CNN; (Note 6) ), a type of deep learning. (DeepInsight method). Such sorting of elements by similarity of image format was found to improve classification performance compared to processing each element individually. In general, however, how to interpret the results obtained from such deep learning models in a biomedical context remained an open question.
Research Details
Professor Tatsuhiko Tsunoda and his colleagues at The University of Tokyo's Graduate School of Science have developed the DeepFeature method, which converts non-image omics data such as genomes into image format and handles them with deep learning (especially CNN), and also uses deep learning to discover which genes and other features should be emphasized. This method starts with image transformation using DeepInsight, followed by a newly developed data compression method (Snowfall) to handle more data elements within a certain pixel frame, classification using the SqueezeNet model, one of CNNs, and a method to visualize regions of interest (CAM) to discover the features of interest, and decoding to the corresponding gene set (Figure 1). In particular, for the part of finding out what is emphasized, a new region aggregation and element decoder (READ) was developed (Figure 2) to find genes and elements with high activity by looking at the activity level of the middle layer of the CNN (Figure 1).
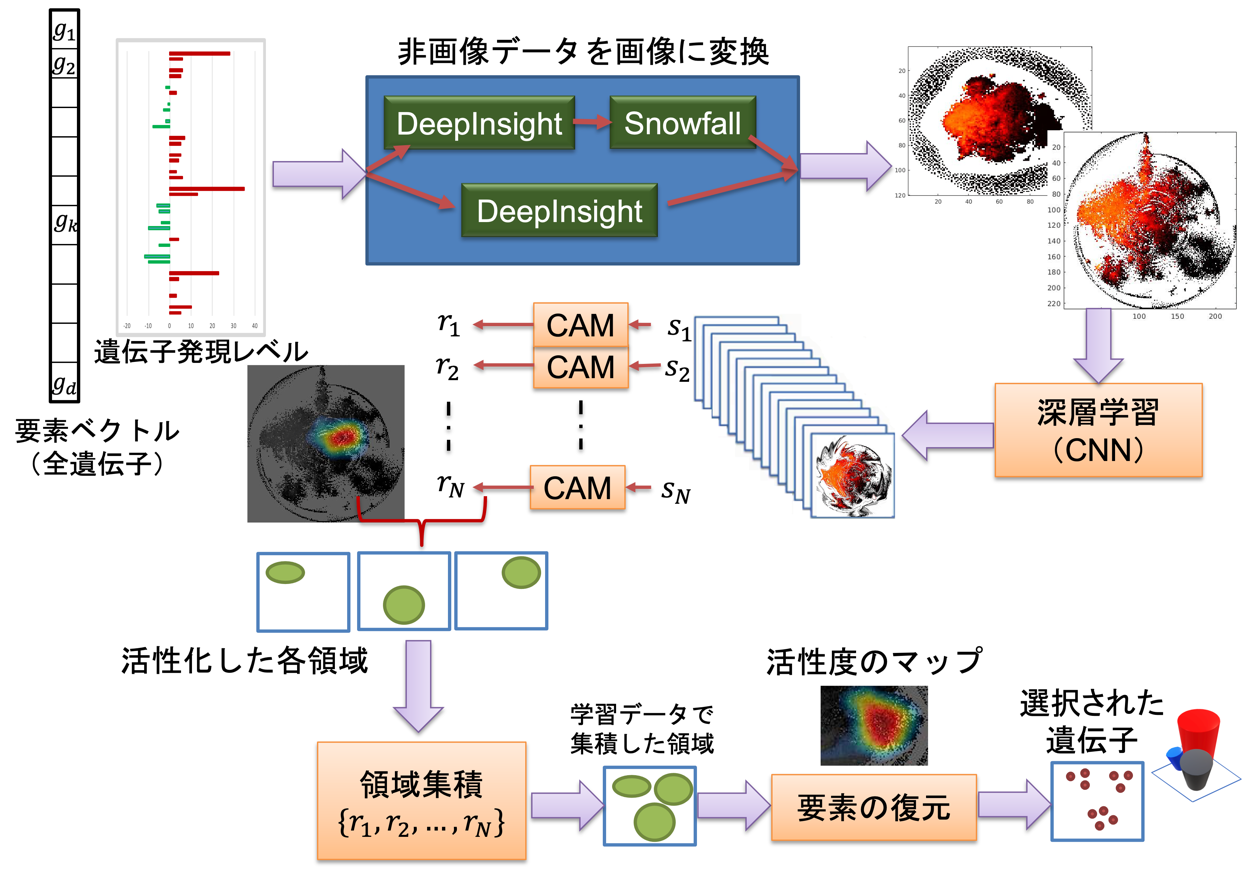
Figure 1: Overall procedure of this method DeepFeature, which converts non-image data, such as genomics, into images and uses convolutional neural networks (CNN) to extract and select features. Modified from this research paper.
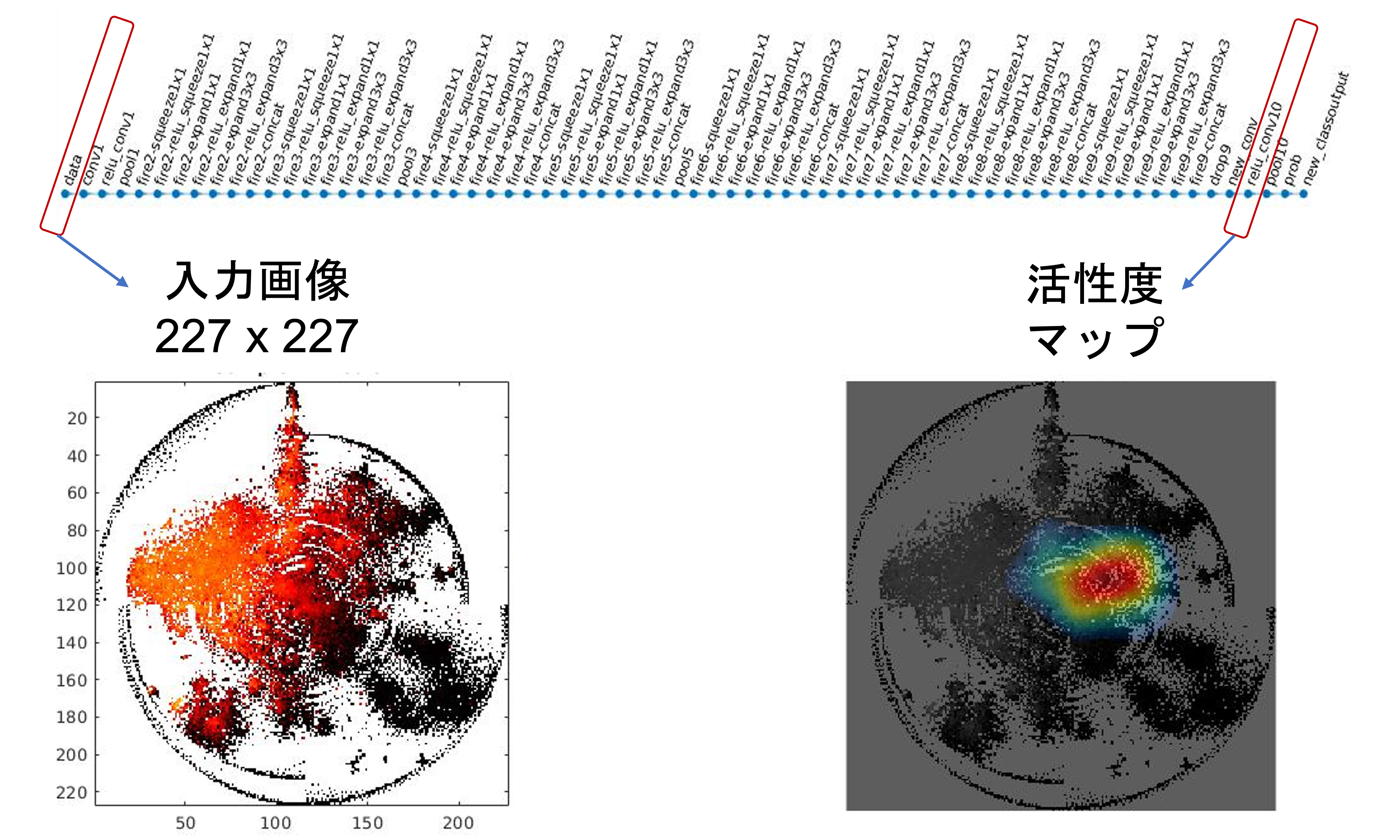
Figure 2: The CNN used internally, the input image sample after converting non-image data into images, and the corresponding activation level of the intermediate layer. Modified from this research paper.
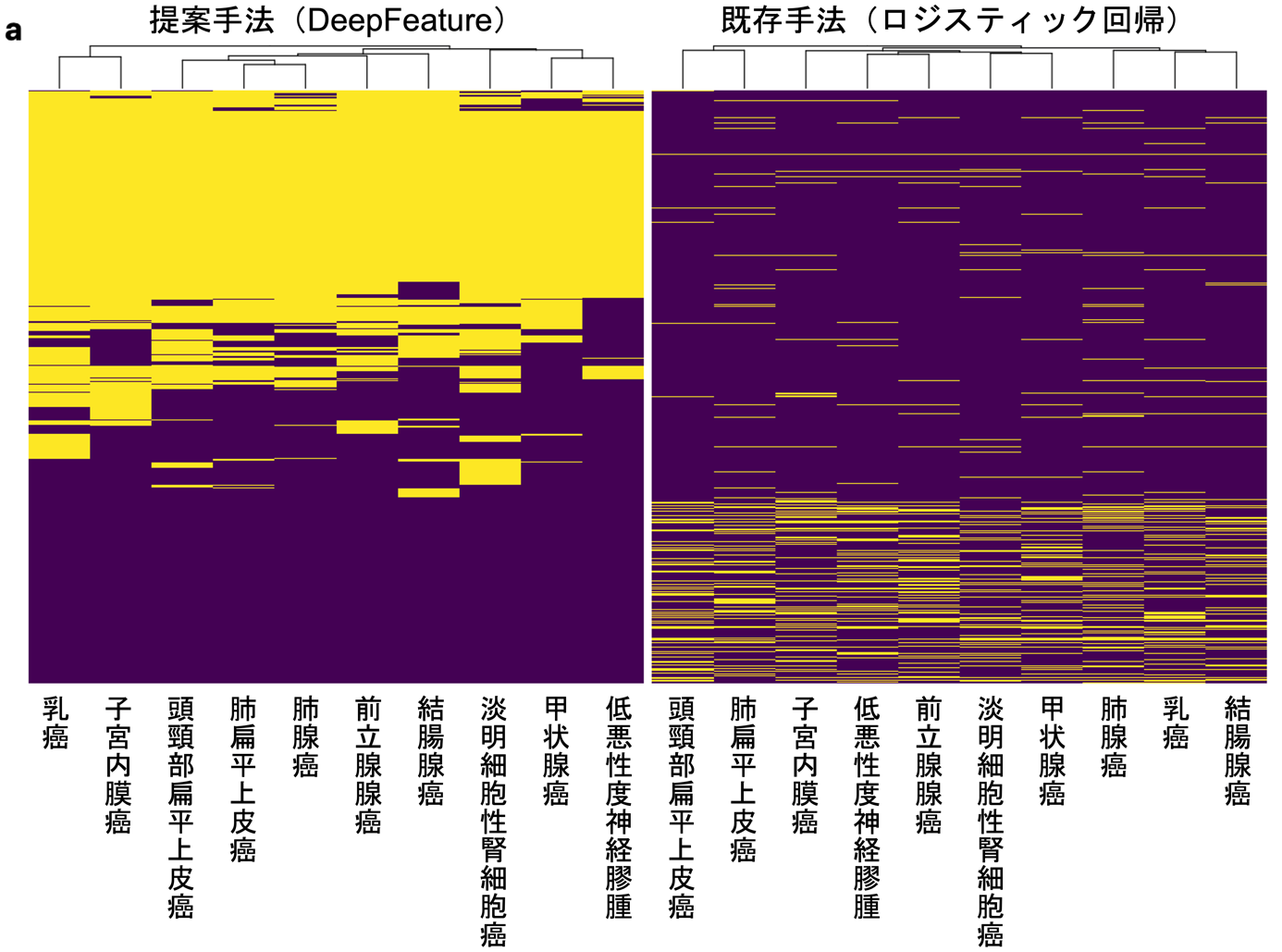
When the DeepFeature method was applied to an experiment to discriminate and predict 10 types of cancer, it was able to identify a set of genes significantly clustered in cancer-related signaling pathways defined in public databases, such as epithelial-mesenchymal transition, coagulation, angiogenesis, hypoxia, and inflammatory response, with excellent prediction performance, The results were found to be much better than other algorithms (Figure 3). Furthermore, when the database was examined for gene set clusters that discriminated each cancer type, novel signaling pathways and biological processes that could discriminate cancer types were also discovered, such as extracellular matrix structure, receptor tyrosine kinase signaling, and GPCR ligand binding.
b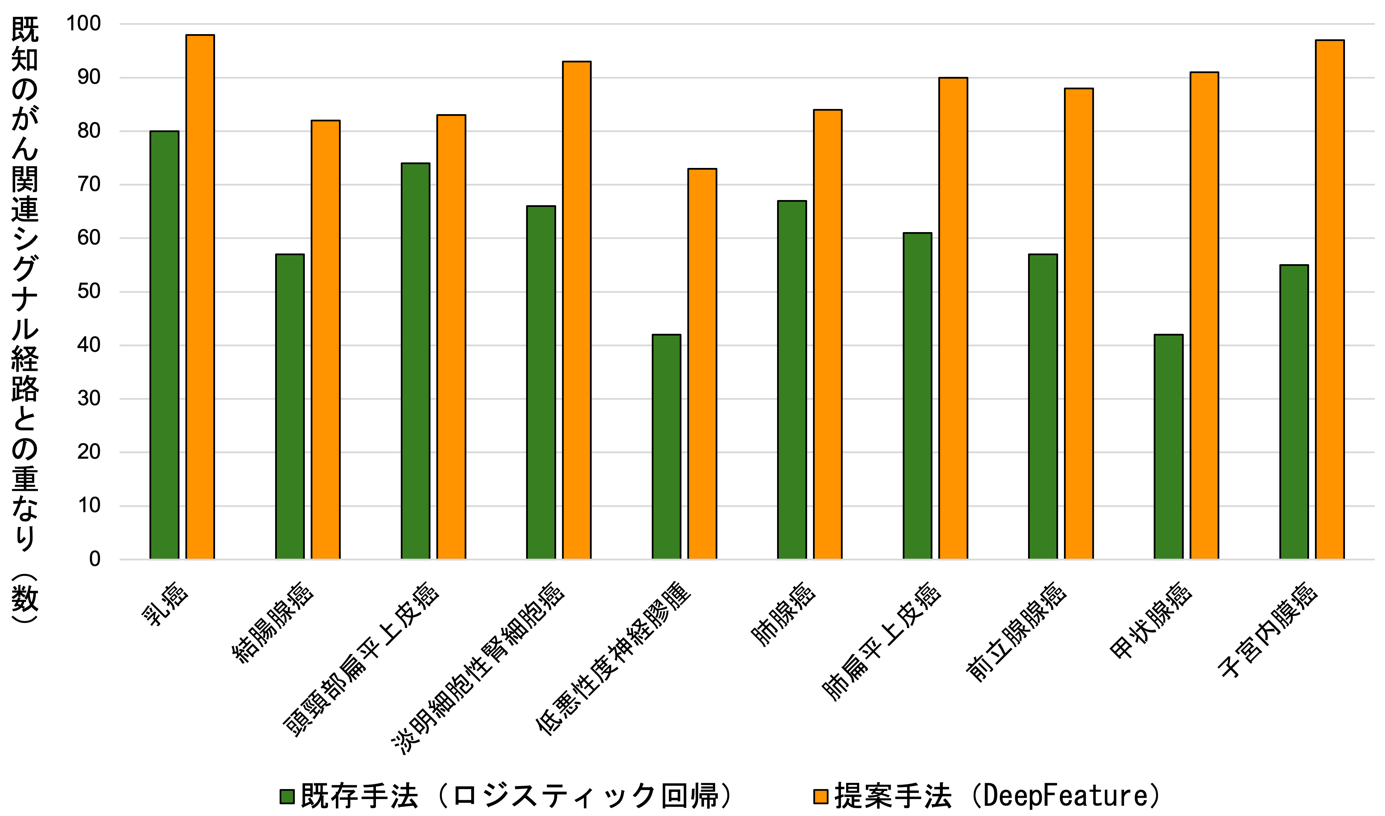
Figure 3: a) Diagram showing the extent to which the genes selected by the DeepFeature and logistic regression methods proposed in this study overlap. Each row represents a gene, and yellow indicates that the gene was selected for that cancer type (column). b) The degree of overlap between the set of genes selected by each method and the set of genes in known cancer-related signaling pathways. The number of signaling pathways that significantly clustered for the genes selected by DeepFeature (orange). For all cancer types, DeepFeature showed higher numbers than logistic regression. Modified from this research paper.
Social Significance, Future Plans, etc.
This is the first proposal in the world for a method that enables biomedical interpretation of what is important in deep learning when genome and other data are imaged and trained by deep learning such as CNN, and extracts important genes and other features. One of the applications is the elucidation of the complex and dynamic mechanism of oncogenesis and overcoming cancer. To elucidate the mechanisms of cancer and discover clinically important subtypes, it is necessary to collect data in a variety of ways, appropriately combine different types of data, and identify meaningful patterns. For this, deep learning-based methods are currently the most flexible and may be able to integrate these different types of data in a more comprehensive manner. And the methodology proposed in this study will enable the effective use of deep learning to discover causal relationships from high-dimensional biomedical data, which can lead to prioritization of diagnostic markers, interpretation of complex multi-omics data, and scientific discovery, in addition to prognosis and treatment effect prediction. Moreover, deep learning, despite its capabilities and many recent advances, is commonly regarded as ultimately producing "black box" models, making it very difficult to discover how the algorithm came to its conclusions. However, with this research as a clue, it will no longer be a dream that deep learning can break out of the so-called "black box" model and discover how it came to its conclusions.
Journal
-
Journal name Briefings in Bioinformatics Title of paper DeepFeature: Feature Selection in non-image data using convolutional neural network Author(s) Alok Sharma*, Artem Lysenko*, Keith A Boroevich, Edwin Vans, Tatsuhiko Tsunoda*, and Tatsuhiko Tsunoda DOI Number 10.1093/bib/bbab297
URL
Terminology
Note 1 Genome
Genome is a composite term from gene (gene) and chromosome (chromosome) and refers to all genetic information in DNA. ↑up
Note 2 Omics
Omics is a field of study that deals with a comprehensive collection of information on biomolecules. Biomolecular information includes, for example, genome, epigenome (total modifications to genome), transcriptome (total gene expression), proteome (total proteins), metabolome (total metabolites), etc. ↑up
Note 3 Multi-omics
A methodology to analyze different omics data such as genome, epigenome, transcriptome, proteome, and metabolome in an integrated manner. Multi-omics data refers to the cohesion of those data. ↑up
Note 4 Deep learning, intermediate layer
Deep learning is a machine learning method using multi-layer neural networks (deep neural networks). Deep Learning. It consists of an input layer that inputs data, multiple intermediate layers that pass on information while performing operations, and an output layer that outputs discrimination results. In this research, the key point is to find out what features deep learning focuses on by looking at the intermediate layers (right side of Figure 2), which are quite close to the output layer. ↑
Note 5 Genomics
A life science field that studies genomes and genes. ↑up
Note 6 Convolutional neural network (CNN)
One of the deep learning methods that demonstrate high performance especially in image classification and discrimination. CNN stands for Convolutional Neural Network. ↑up


