DATE2021.06.08 #Press Releases
The search for molecular mechanisms of stable and economical heart contractions planted in cardiac myosin
Overview of the press release
Researchers in the Graduate School of Science and their collaborators have discovered that cardiac myosin has unique properties, and that these properties are key to facilitating efficient heart contractions, contributing to stable blood flow followed by rapid relaxation, and saving energy expenditure.
Since the molecular structure of cardiac myosin is very similar to that of skeletal muscle myosin, it has been generally imagined that the contraction mechanism of the heart is similar to that of skeletal muscle. However, in the heart, periodic stable contraction followed by rapid relaxation is important, while skeletal muscle contraction requires an extremely wide range of contraction forces and velocities, from small forces to explosively large forces, and from slow to fast contractions, depending on external demands. Therefore, the research team expected that the molecular mechanism of contraction should be different between the heart and muscle.
The researchers characterized and compared the molecular properties between cardiac and skeletal myosins using single molecule measurement techniques, based on the idea that the molecular function specific to stable and efficient heart contractions, a unique property of cardiac myosin, is not present in skeletal myosin. They found that the force generation of the cardiac myosin ensemble was different from that of the skeletal muscle myosin ensemble. To clarify how this difference contributes to cardiac contraction, they conducted another experiment to evaluate the dynamics of single myosins and a developed simulation model. Their findings suggest that the molecular properties of cardiac myosin are very important for heart function (Figure 1).
“The fact that we were able to link the function of a single myosin molecule to the function of an organ composed of billions of myosin molecules by conducting a comprehensive study that combined the two experiments and the simulation was very exciting and brought us closer to our research goal,” said Motoshi Kaya, an assistant professor in the Department of Physics, Graduate School of Science.
The main cause of hypertrophic cardiomyopathy, which is also a cause of sudden death, is mutations in contractile proteins. Nearly half of these mutations originate in cardiac myosin. Therefore, by comparing the properties of wild-type cardiac myosin with those of mutant myosins, the researchers hope to provide valuable information to elucidate the pathogenesis of hypertrophic cardiomyopathy and develop therapeutic agents. In addition, since the genes of cardiac myosin and slow muscle myosin are identical, the molecular mechanism of the heart contractions discovered in this study may be useful to understanding the contraction mechanism of slow muscles.
“I believe that there are no other organs with such periodic and regular structures as skeletal muscles and the heart, and that such structures are physiologically meaningful,” said Kaya. “In particular, I am motivated to understand whether spontaneous functions, which emerge when proteins are grouped together, are responsible for the complex and highly functional contractions of muscles and the heart.”
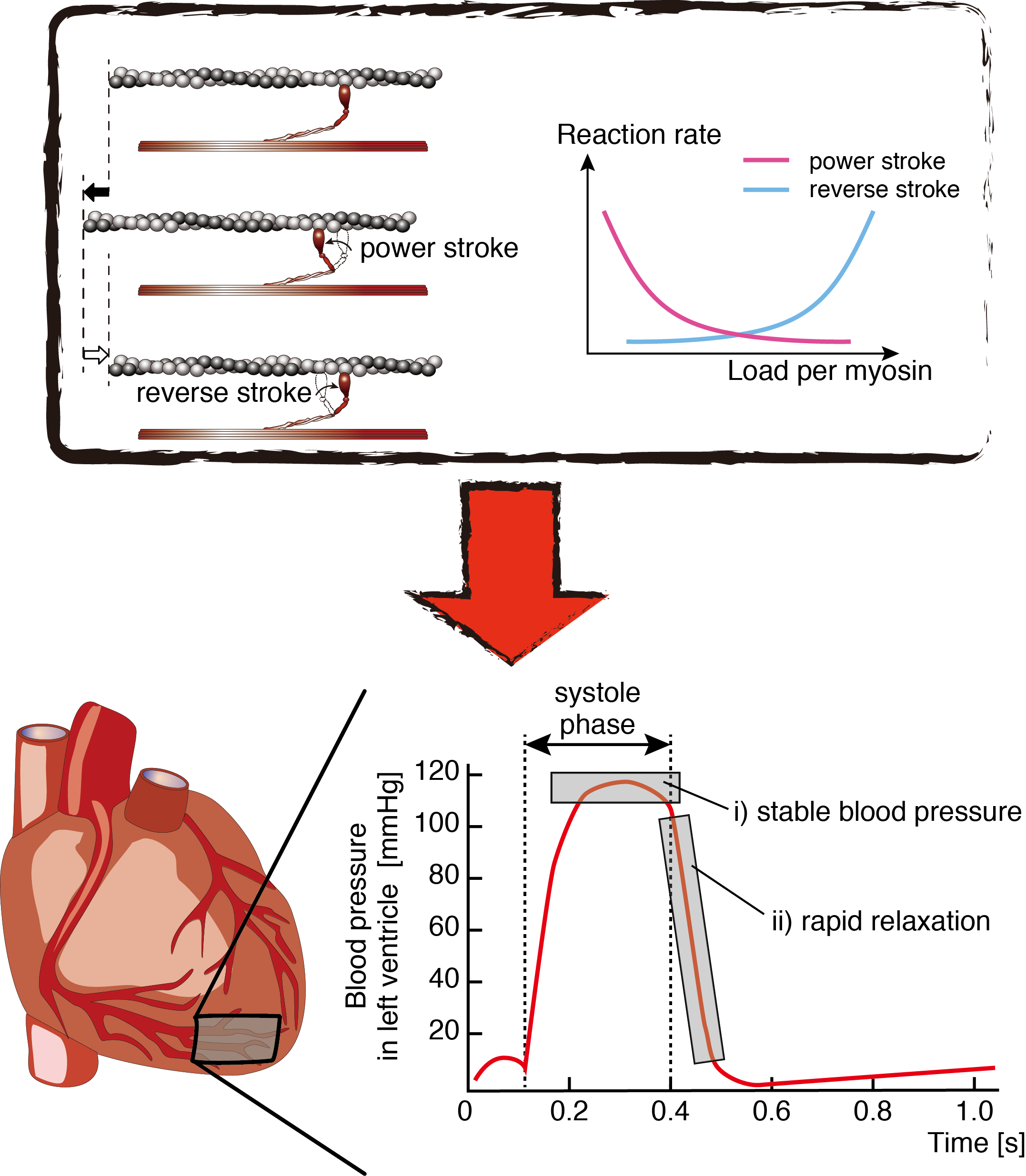 Figure 1: Unique molecular properties of cardiac myosin for efficient heart contractions. Highly load-dependent reaction rates of power stroke and reverse stroke was found to be the unique molecular property of cardiac myosin (top) and is key to facilitating efficient heart contractions, such as the maintenance of stable systolic pressure and the rapid relaxation of pressure in the latter systole phase (bottom).
Figure 1: Unique molecular properties of cardiac myosin for efficient heart contractions. Highly load-dependent reaction rates of power stroke and reverse stroke was found to be the unique molecular property of cardiac myosin (top) and is key to facilitating efficient heart contractions, such as the maintenance of stable systolic pressure and the rapid relaxation of pressure in the latter systole phase (bottom).Publication details
| Journal |
Proceedings of the National Academy of Sciences of the United States of America (PNAS)
|
|---|---|
| Title |
A reverse stroke characterizes the force generation of cardiac myofilaments, leading to an understanding of heart function.
|
| Authors |
Yongtae Hwang, Takumi Washio, Toshiaki Hisada, Hideo Higuchi* and Motoshi Kaya*
|
| DOI | 10.1073/pnas.2011659118 |
| URL | https://www.pnas.org/content/118/23/e2011659118 |


