DATE2022.01.24 #Press Releases
Successful demonstration of the evolutionary process of the operon structure
Disclaimer: machine translated by DeepL which may contain errors.
Yuki Kanai (Doctoral Student, Department of Biological Sciences)
Sanyoshi Tsuru, Project Assistant Professor, Universal Biology Institute
Chikara Furusawa, Professor, Universal Biology Institute
Key points of the presentation
- We proposed a new hypothesis that the operon (Note 1), the basic structure of the prokaryotic genome, evolved through the function of the insertion sequence (Note 2), and succeeded in a proof experiment.
- For the first time in the world, the evolutionary process of the formation of prokaryotic operons has been observed in a way that can be reproduced in the laboratory.
- The ability to repeatedly observe the evolution of operons is expected to lead to a better understanding of how gene regulation mechanisms have evolved and become more complex.
Summary of Presentation
In prokaryotes, multiple genes with related functions are regulated by grouping them into structures called operons. The origin of the elaborate operons of prokaryotes has long been a mystery.
In this study, Graduate Student Yuki Kanai, Assistant Professor Mitsuyoshi Tsuru, Professor Chikara Furusawa, and their colleagues at the Graduate School of Science, The University of Tokyo, have proposed a new hypothesis that the evolution of operon formation is driven by sequences called insertion sequences, which are universal in prokaryotic genomes. By culturing E. coli in the laboratory under conditions of high insertion sequence activity, they demonstrated that operons can be formed as hypothesized. This is the first study to demonstrate one of the previously unknown mechanisms of prokaryotic operon formation by observing the evolutionary process.
The fact that operons can be formed by insertion sequences commonly found in pathogenic bacteria such as pathogenic Escherichia coli O157 provides new insights into the acquisition and control of bacterial virulence. This study has also revealed experimental conditions under which operon formation can be reproduced repeatedly. These results are expected to elucidate the process by which primitive organisms acquired operons and approached the current prokaryotes.
Publication details
Background of the research Problems in previous studies
An operon is a structure in which multiple genes on the genomic DNA are placed under a single promoter (Note 3 ). When functionally related genes are integrated into an operon, multiple genes can be regulated simultaneously according to the environment.
The rationale for the structure of an operon has been examined from various perspectives. However, the mechanism by which multiple genes at distant locations on DNA come together to form an operon during the process of evolution by random mutation was not understood (Figure 1).
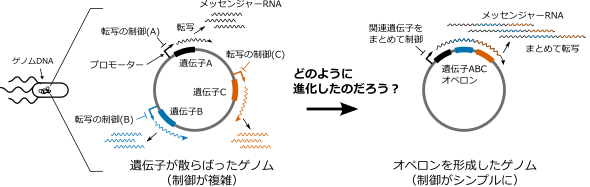
Figure 1: Purpose of the study. We know that forming an operon is rational because it facilitates the control of transcription. How operons evolve, however, has remained a mystery.
Research Details
Insertion sequences (Fig. 2-a) are sequences that encode a single gene, the transposase gene, but have the ability to cause various mutations such as insertion, deletion, and excision in other sequences (Fig. 2-b), and these individual mutations have been frequently observed. The IDE (Insertion, Deletion, Excision) model of operon formation proposed in this study proposes that the combination of these mutations caused by insertion sequences brings multiple genes located far apart on the genomic DNA closer together and drives the evolution of operon formation ( The IDE hypothesis (see Figure 2-c for details).

Figure 2: Evolutionary process of operon formation by insertion sequences. (A) Structure of insertion sequence. (b) Three functions of insertion sequences. The insertion sequence is depicted in simplified form as a rectangle. (c) Schematic diagram of the evolutionary process based on operon formation using the IDE model. Genes are depicted as bold arrows. An operon is formed in three steps. (i) An insertion sequence is inserted between two genes (black and orange). (ii) The insertion sequence brings the two genes closer together by excising the surrounding DNA at a high frequency. Three: The insertion sequence is excised and the operon is formed.
To verify the proposed hypothesis, the following experiments were conducted. First, we genetically engineered E. coli with a DNA sequence in which the insertion sequence was "inserted" between the antibiotic resistance gene (Note 5 ) and the two red and yellow fluorescent protein genes (Note 6) (Figure 3-a, DNA sequence of the original E. coli). By culturing this E. coli in a medium containing antibiotics, we expected that the antibiotic resistance gene and the red fluorescent protein gene would form a new operon, and a mutant E. coli showing stronger red fluorescence would emerge. In addition, for operon formation by the IDE model, it is essential that the resection and truncation functions of inserted sequences produce diverse genotypes. Therefore, by targeting the formation of fluorescent protein gene operons, we were able to measure the fluorescence intensity of E. coli at high speed in each cell, allowing us to observe the genetic diversity caused by the activity of insertion sequences.

Figure 3: Demonstration of operon formation of antibiotic resistance genes and red fluorescent protein genes. (a) Genotypic changes before and after operon formation by the IDE model. The activity of the insertion sequence brought the antibiotic resistance gene and the red fluorescent protein gene closer together and formed an operon. (b) Fluorescence intensity per cell in E. coli (left: without insertion sequence activity; right: with activity). The distance between the fluorescent protein gene and the surrounding promoter changes depending on the excision and truncation activity of the insertion sequence, showing a variety of fluorescence intensities. (c) Messenger RNA was detected by RT-PCR method(Note 8 ). (-) indicates the conditions used to confirm that no false positives were produced. (d) We confirmed that the expression level of the red fluorescent protein gene can now be controlled via the promoter of the antibiotic resistance gene (whose transcriptional activity switches in response to drug addition).
When E. coli were incubated in the laboratory for one day (less than 20 generations ), mutants with diverse fluorescence were generated under conditions of high activity of the insertion sequence (Figure 3-a). To find the mutants that formed the operon among them and to determine their genotype, some cells were aliquoted with the fluorescent cell sorter (Note 7). The genotypes of the aliquoted cells were examined by DNA sequencing, and it was found that the mutant exhibited operon formation based on the IDE model (Figure 3-a). We also confirmed that messenger RNAs spanning two genes were generated in this mutant (Fig. 3-c) and that transcription of the two genes was simultaneously regulated (Fig. 3-d). This result implies that these genes have operon properties. The appearance of such mutants depending on the activity of the insertion sequence demonstrates that an operon can be formed by the IDE model.
Future Developments
Insertion sequences are highly active in many pathogenic bacteria, not only E. coli. Therefore, the hypothesis and experimental evidence that insertion sequences can form operons provide new insights into how pathogenic bacteria acquire operons that are responsible for their characteristics such as drug resistance and virulence.
Insertion sequences are simple genes, and primitive cells may have had similar genes. In addition, the various mutations caused by insertion sequences are closely related to the mechanism of self-replication of the insertion sequence itself. Therefore, this study suggests that organisms may have formed operons and acquired sophisticated gene regulation through the process of symbiosis with "selfish" genes such as insertion sequences.
Furthermore, through empirical experiments, we have clarified the conditions under which the formation process of an operon driven by an insertion sequence can be reproduced repeatedly in the laboratory. Further development of this study to explore the conditions under which operons form at a sufficient rate in primordial cells is expected to lead to an understanding of how life has acquired complex gene regulatory mechanisms.
Acknowledgments
This work was supported by Grant-in-Aid for Scientific Research (21J20693, 18H02427, 17H06389) from the Japan Society for the Promotion of Science (JSPS), Japan Science and Technology Agency (JST) and ERATO (JPMJER1902).
Journals
-
Journal name Nucleic Acids Research Title of paper Experimental demonstration of operon formation catalyzed by insertion sequenceAuthor(s) Yuki Kanai, Saburo Tsuru*, Chikara Furusawa*, Chikara FurusawaDOI number https://doi.org/10.1093/nar/gkac004
Terminology
Note 1 Operon
A gene written as DNA sequence information is transcribed from a promoter sequence to messenger RNA, which is further translated into a protein to perform its function. The structure of a DNA sequence in which multiple genes are transcribed from a single promoter sequence to messenger RNA is called an operon. By grouping genes into operons, a group of functionally related genes can be regulated simultaneously (Figure 1). ↑up
Note 2 Insertion sequence
A generic term for genes with a simple structure that replicates independently of the genome, which is universally found in prokaryotes. An insertion sequence consists only of a transposase gene (and its related genes), which is an enzyme for its own self-replication, and a recognition sequence that defines its own boundaries (Fig. 2-a). ↑up
Note 3 Promoter
A sequence of DNA used by RNA polymerase, the enzyme that makes messenger RNA, as a marker to initiate transcription. ↑up
Note 4 Operon theory
A hypothesis on gene regulation in organisms proposed by Jacob and Monot in 1961. They predicted the basic principle of the regulation of gene expression in organisms and received the Nobel Prize in Physiology or Medicine in 1965 for their "discoveries concerning the genetic control of enzymes and viral synthesis. They focused on the phenomenon that E. coli regulates the amount of enzymes involved in lactose metabolism in response to the presence or absence of lactose in the culture medium, and analyzed lactose-metabolizing mutants of E. coli. From this, they found that E. coli can control lactose metabolism because it has a gene (repressor) that inhibits the synthesis of the "messenger" (later messenger RNA ) of genes that have enzymatic activity required for lactose metabolism, and a gene that is recognized by the repressor and used to suppress the expression of downstream genes ( operator) because E. coli has them. Assuming the existence of repressors and operators, he named the genetic unit of co-ordinate expression (" operon"), which is simultaneously regulated by them. He predicted that the group of genes arranged collectively in the genome is exactly this operon. ↑up
Note 5: Antibiotic resistance gene
Bacteria expressing this gene can grow on medium with antibiotics added. ↑up
Note 6 Fluorescent protein gene
Cells expressing this gene become fluorescent, and gene expression can be examined while the cells are alive. A fluorescent protein that glows in various colors has been developed by modifying the green fluorescent protein of the oneon jellyfish. ↑up
Note 7 Fluorescent cell sorter
A device in which cells are passed one by one through a narrow water stream, fluorescence is measured, and cells are sorted according to fluorescence intensity. ↑↑
Note 8 RT-PCR method
RNA is converted to DNA (reverse transcription), and the resulting DNA is amplified by PCR for detection. ↑↑


