DATE2022.02.05 #Press Releases
Quantitative understanding of metabolic changes and regulation in obesity using mathematical models
Disclaimer: machine translated by DeepL which may contain errors.
Saori Uematsu (Graduate School of Frontier Sciences, 3rd Year Doctoral Student)
Satoshi Ohno (Assistant Professor, Molecular Genetics Research Laboratory)
Shinya Kuroda (Professor, Department of Biological Sciences)
Key points of the presentation
- We developed a mathematical model OMELET to analyze metabolic changes and their regulation from multi-omics data (Note 2 ) without using isotope labeling experiments (Note 1).
- Using OMELET, we quantitatively analyzed changes in liver glucose metabolism and the regulation that causes these changes in an obese mouse model.
- Since OMELET is applicable to any species and any metabolic pathway, it is expected to contribute to the elucidation of the pathogenesis of various metabolic diseases as well as obesity.
Summary of Presentation
Saori Uematsu, Doctoral student, Graduate School of Frontier Sciences, The University of Tokyo, Satoshi Ohno, Assistant Professor, Graduate School of Science, The University of Tokyo, and Shinya Kuroda, Professor, Graduate School of Medicine, Kyushu University, Masaki Matsumoto, Professor, Niigata University Graduate School of Medical and Dental Sciences, Atsushi Hatano, Assistant Professor, The University of Tokyo, Professor Minoru Suzuki, Graduate School of Professor Minoru Suzuki, Graduate School of Mathematical Sciences, Niigata University, Associate Professor Tomoyoshi Soga and Project Associate Professor Akiyoshi Hirayama, Institute for Quantitative Biosciences, Keio University, and others, we have quantitatively analyzed changes in liver glucose metabolism and its regulation in obesity using our originally developed mathematical model OMELET.
The liver plays a central role in maintaining systemic glucose homeostasis, and abnormalities in liver glucose metabolism are observed in obesity and diabetes. Until now, isotope labeling experiments have been used to measure abnormal reaction rates (metabolic fluxes) of metabolic reactions, but this has been costly and time-consuming. In addition, each metabolic reaction is regulated by various molecules such as enzymes and metabolites, but it was not well understood which molecules and to what extent they affect metabolic abnormalities.
In this study, the research group developed OMELET, a mathematical model to estimate metabolic flux from multi-omics data, and analyzed both metabolic flux and its regulation in mouse liver without using isotope labeling experiments to quantitatively clarify changes in liver glucose metabolism and its causes in a mouse model of obesity. This study clarified the metabolic fluxes and regulation of liver glucose metabolism in obese mouse models. Although metabolic fluxes of both glycogenesis and its rate-limiting reaction, phosphoenolpyruvate carboxykinase (PEPCK) (Note 3) reaction, were increased in obesity, the increase in metabolic fluxes in glycogenesis was mainly caused by transcripts, whereas in PEPCK, metabolic fluxes were caused by substrate metabolites PEPCK, however, the increase in metabolic flux was mainly driven by substrate metabolites.
The OMELET developed by the research group is applicable to any species and metabolic pathway, and is expected to contribute to the elucidation of the pathogenesis of various metabolic diseases as well as obesity.
Publication details
Background of Research
Glucose (glucose) is one of the most important nutritional sources for living organisms, and the human body is equipped with homeostatic mechanisms to maintain a constant blood glucose level (blood glucose concentration). The liver is a major organ for glucose homeostasis, and contributes to the maintenance of blood glucose levels by producing glucose and releasing it into the blood via glycogenolysis and glycogenesis during fasting. However, in obesity and diabetes, blood glucose levels are chronically high and the glucose homeostasis mechanism is disrupted. Therefore, investigation of liver glucose metabolism is important for understanding the pathogenesis of obesity and diabetes.
To understand metabolic abnormalities in obesity, we need quantitative information on how metabolism is altered and to what extent multiple regulatory factors, such as enzymes and metabolites, each contribute to these changes. To directly understand metabolic changes, it is necessary to measure the reaction rate of metabolic reactions (metabolic flux). Metabolic fluxes are generally measured by isotope labeling experiments, but measuring metabolic fluxes, especially in specific organs in vivo, has been costly and time-consuming. For example, it is necessary to optimize experimental conditions, such as the dose of isotope-labeled metabolite to be administered and the sampling time, and analysis that takes into account metabolism in organs other than the target organ is also necessary. In addition, although recent developments in measuring instruments have made it possible to measure the regulatory factors involved in metabolic flux changes on a large scale, the extent to which each regulatory factor influences metabolic changes was not yet known.
Therefore, this research group developed a mathematical model to analyze metabolic flux and its regulation using multi-omics data without isotope labeling experiments, with the aim of quantitatively understanding abnormal liver glucose metabolism in obesity.
Research Details
The research group developed Omics-Based Metabolic Flux Estimation without Labeling for Extended Trans-omic Analysis (OMELET), a method for estimating both metabolic flux and its regulation from multi-omics data (Figure 1). OMELET is based on Bayesian inference ( ) and estimates metabolic fluxes and reaction rate parameters using multi-omics data on metabolite, enzyme protein, and transcript levels and information on target metabolic pathways as input. Reaction rate parameters are used to calculate the contribution of regulatory factors to differences in metabolic fluxes between conditions.
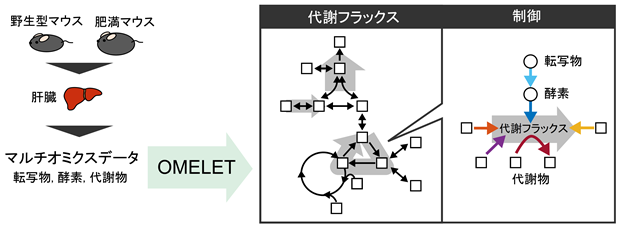
Figure 1: The mathematical model OMELET, which quantitatively analyzes metabolic changes and their regulation from multi-omics data, was used to analyze abnormal liver glucose metabolism in obesity.
Estimation of metabolic fluxes of liver glucose metabolism
The research group obtained multi-omics data on metabolites, enzyme proteins, and transcripts from the livers of fasting wild-type and ob/ob mouse models of obesity and diabetes. Many metabolites, enzyme proteins, and transcripts belonging to glucose metabolism were increased in obesity. However, it was not clear how metabolic fluxes are altered in obesity only from the multi-omics data. We first applied OMELET to these multi-omics data to estimate metabolic fluxes of liver glucose metabolism and quantify changes in metabolic fluxes associated with obesity. This confirmed that OMELET can estimate metabolic fluxes accurately enough without isotope labeling experiments. OMELET also showed that metabolic fluxes of gluconeogenesis and pyruvate cycle are greatly increased in obese mice (Figure 2).
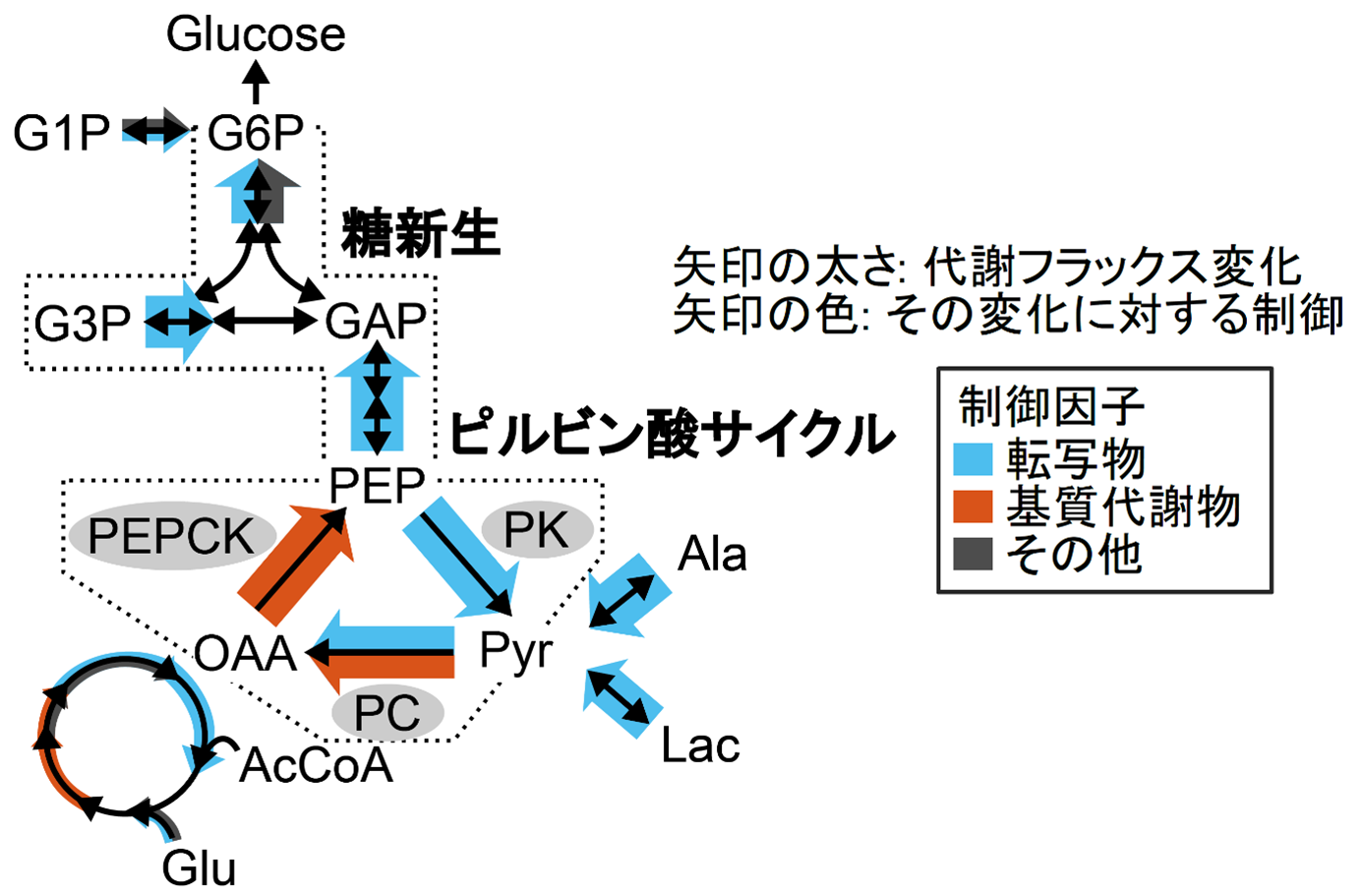
Figure 2: Liver glucose metabolic changes in obesity and the main controls on these changes. The increased metabolic flux of gluconeogenesis in obese mice was mainly caused by increased transcripts, not metabolites. In contrast, the increased metabolic flux in the pyruvate cycle was first caused by increased PK flux due to increased PK transcripts, which led to increased downstream PC and PEPCK substrate metabolites, and finally increased PEPCK flux driven by increased substrate metabolites.
Contribution of Obesity to Metabolic Flux Changes in Obesity
To determine the extent to which each regulatory factor, such as enzymes and metabolites, affected the metabolic flux changes caused by obesity, we calculated the contribution ratio of each regulatory factor to the metabolic flux changes associated with obesity from the multi-omics data and the reaction rate parameters estimated from OMELET. The results showed that the increased metabolic flux of glycogenesis in obese mice is not caused by metabolites, but mainly by increased transcripts via enzyme expression (Figure 2). On the other hand, the mechanism of increased pyruvate cycle flux differs by reaction, with pyruvate kinase (PK) (Note 5 ) primarily driven by transcript levels, pyruvate carboxylase (PC) (Note 6) by both transcript and substrate metabolites, and phosphoenolpyruvate carboxylase (PEPCK) primarily by substrate metabolites, and that it is caused by an increase in This suggests that the mechanism of increased metabolic flux in the pyruvate cycle is that increased PK flux is first caused by increased PK transcripts, which leads to increased downstream PC and PEPCK substrate metabolites, and finally increased PEPCK flux is driven by increased substrate metabolites The results suggest a mechanism whereby the increase in PEPCK flux is driven by the increase in substrate metabolites.
In summary, by integrating the contribution of regulatory factors to metabolic fluxes and metabolic fluxes estimated using the multi-omics data OMELET, a quantitative trans-omics network (Note 7), we have revealed some aspects of obesity-induced changes in liver glucose metabolism and the regulation that causes these changes. The results are shown in the following table.
Social Significance and Future Plans
In recent years, multi-omics data have been accumulated for various species of organisms through the development of omics measurement technology. On the other hand, it is now a challenge to integrate multi-omics data and derive useful biological findings, and the need for the development of new analysis techniques is increasing. The OMELET developed by this research group is applicable to any species and metabolic pathways for which multi-omics data can be obtained, and is expected to contribute to the elucidation of the pathogenesis of various metabolic diseases in mice and humans, for which isotope labeling experiments are difficult.
This research was supported by Grant-in-Aid for Scientific Research on Innovative Areas (proposed research area) "Metabolic Adaptation of Type 2 Diabetes Mellitus" (project number: JP17H06299, JP17H06300, PI: Shinya Kuroda) from the Japan Society for the Promotion of Science and Technology (JSPS), by the Strategic Creative Research Promotion Program "Multicellular Spatiotemporal Interactions" from the Japan Science and Technology Agency (JST), and by the Japan Science and Technology Agency (JST). The project was funded by the Japan Science and Technology Agency as part of the Strategic Creative Research Promotion Program "Creation of a Quantitative Analysis Platform for Understanding Spatio-Temporal Interactions among Multicellular Organelles" under the research theme "Elucidation of Metabolic Regulation in Multicellular Organelles Using Space-Time Transomics" (project title: JPMJCR2123; PI: Shinya Kuroda).
Journal
-
Journal name iScienceTitle of paper Multi omics based label free metabolic flux inference reveals obesity associated dysregulatory mechanisms in liver glucose metabolismAuthor(s) Saori Uematsu, Satoshi Ohno*, Kaori Y. Tanaka, Atsushi Hatano, Toshiya Kokaji, Yuki Ito, Hiroyuki Kubota, Ken-ichi Hironaka, Yutaka Suzuki, Masaki Matsumoto, Keiichi I. Nakayama, Akiyoshi Hirayama, Tomoyoshi Soga, Shinya Kuroda*, and Shinya KurodaDOI Number https://doi.org/10.1016/j.isci.2022.103787
Terminology
1 Isotope labeling experiments
An experimental method to estimate the metabolic state of a metabolite labeled with a stable isotope by inducing the metabolite to be taken up into a cell or an individual and tracking its diffusion and changes. ↑up
Note 2 Multi-omics data
Multiple omics data of different types of metabolites, enzyme proteins, transcripts, etc. ↑↑
Note 3 Phosphoenolpyruvate carboxykinase (PEPCK)
A reaction that belongs to the pyruvate cycle and converts oxaloacetate to phosphoenolpyruvate. It is also one of the rate-limiting reactions of the glycogenic pathway. ↑up
Note 4 Bayesian estimation
A statistical method that estimates parameters as a distribution that reflects the variability of the observed data and parameters themselves, rather than the value that maximizes the probability of generating the observed data. ↑up
Note 5 Pyruvate kinase (PK)
A reaction that converts phosphoenolpyruvate to pyruvate, a member of the pyruvate cycle. It is one of the rate-limiting reactions in the glycolytic system. ↑up
Note 6 Pyruvate carboxylase (PC)
A reaction that converts pyruvate to oxaloacetate, belonging to the pyruvate cycle. One of the replenishment pathways to the citric acid circuit. ↑up
Note 7: Transomics network.
A large-scale metabolic regulatory network spanning multiple omics hierarchies of transcripts, proteins, and metabolites. ↑


