DATE2023.01.13 #Press Releases
Elucidating the mechanism of heterochromatinization of DNA with repeated sequences
Disclaimer: machine translated by DeepL which may contain errors.
~Proposing a new model to answer a 40-year-old mystery~.
Hokkaido University
Graduate School of Science, The University of Tokyo
Research Center for Advanced Science and Technology, The University of Tokyo
Summary of Presentations
A research group led by Professor Yota Murakami and Academic Support Staff Takahiro Asanuma at the Graduate School of Science, Hokkaido University, Associate Professor Soichi Inagaki and Professor Tetsuhito Kakutani at the Graduate School of Science, The University of Tokyo, and Senior Research Fellow Hiroyuki Yutani at the Research Center for Advanced Science and Technology, The University of Tokyo, has succeeded in clarifying one aspect of the mechanism by which repeated sequences of DNA are selectively The research group has succeeded in clarifying one aspect of the mechanism by which repeated DNA sequences are selectively heterochromatinized in eukaryotic cells.
Eukaryotic DNA is contained within the nucleus in a bead-like structure wrapped around proteins called histones. This structure is called chromatin, and is broadly classified into euchromatin and heterochromatin based on its properties. In euchromatin, genes on DNA are actively expressed, while in heterochromatin, gene expression is forcibly repressed. These two contrasting states are determined by the chemical modifications of the histones around which the DNA is wrapped; for example, heterochromatin is known to be characterized by its methylation modification (H3K9me modification). Such regulation of gene expression via chromatin state is involved in various biological phenomena and is considered to be one of the fundamental mechanisms of eukaryotic cells.
Interestingly, since the 1980s, during the course of this research, it has been noted that heterochromatin tends to form in DNA regions that have been repeatedly sequenced. This tendency is not limited to repeats with a specific base sequence as the basic unit, but is also observed when entire genes are duplicated and repeated. These facts suggest that eukaryotic cells somehow recognize the "repeated" feature of DNA sequences and promote heterochromatin formation in these regions. However, the mechanism of this mechanism has not yet been clarified.
In this study, the research group artificially reproduced this phenomenon using fission yeast, a model organism, and found that a protein that originally removes heterochromatin (i.e., H3K9me modification) conversely promotes heterochromatin formation (i.e., H3K9me modification) via a mechanism called RNAi in repeated DNA sequences. The results revealed that the protein plays two opposing roles, promoting heterochromatin formation (i.e., H3K9me modification) via the mechanism of RNAi in repeated DNA sequences. This result proposes a new model to answer the previous question of why heterochromatin is selectively formed by repetitive DNA sequences.
Heterochromatin formation by repeated sequences is involved in various biological phenomena regardless of species, and it is known, for example, that some diseases in humans are caused by such defects. Therefore, elucidation of the mechanism will be useful not only for understanding the fundamental mechanism of eukaryotic cells, but also for searching for a cure for human diseases in the future.
The research results were published online in Genes & Development on Tuesday, December 20, 2022.
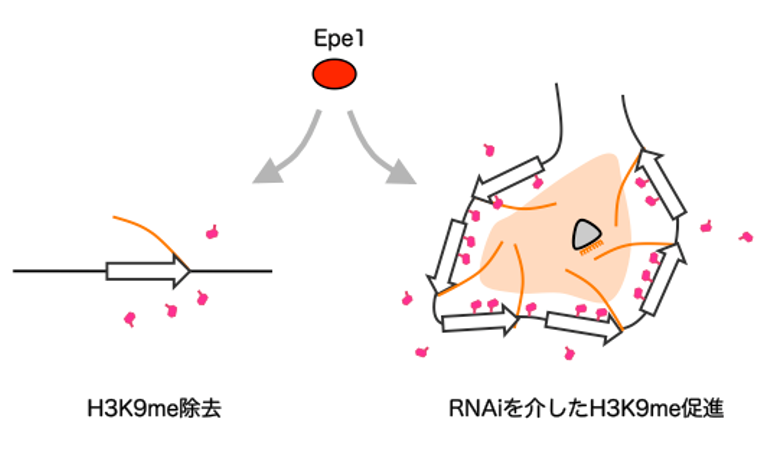
Figure: Epe1, an H3K9me removal factor, has two opposing roles
For more information, please visit the Hokkaido University website.


