DATE2022.11.04 #Press Releases
Mechanisms for coping with ribosome collisions in mammals
Disclaimer: machine translated by DeepL which may contain errors.
-Ubiquitin modification-mediated elimination of translational stagnation reproduced in vitro
The University of Tokyo Institute of Medical Science
Graduate School of Science, The University of Tokyo
Summary of presentation
When ribosomes are strongly stalled during translation, they collide with subsequent ribosomes. Defects in RQC have been shown to be associated with shortened lifespan and neurodegenerative diseases in model organisms. The RQC deficiency has been shown to be associated with shortened lifespan and neurodegenerative diseases in model organisms.
In this study, Professor Toshifumi Inada of the Department of RNA Regulation, Institute of Medical Science, Graduate School of Science and Department of Computational Biology and Medical Sciences, Graduate School of Pharmaceutical Sciences, Tohoku University, graduate student Momoko Narita, and Roland Beckmann of the Gene Center, University of Munich, and their research group have investigated the RQC in yeast cells, mainly in the presence of yeast germination. In their previous studies, mainly using budding yeast, the research group has reported that RQC occurs by the molecular mechanisms of (1) ubiquitination of collision ribosomes, (2) ribosomal subunit dissociation, and (3) degradation of defective proteins in the process of synthesis. However, the molecular mechanism of RQC in mammals remains unclear.
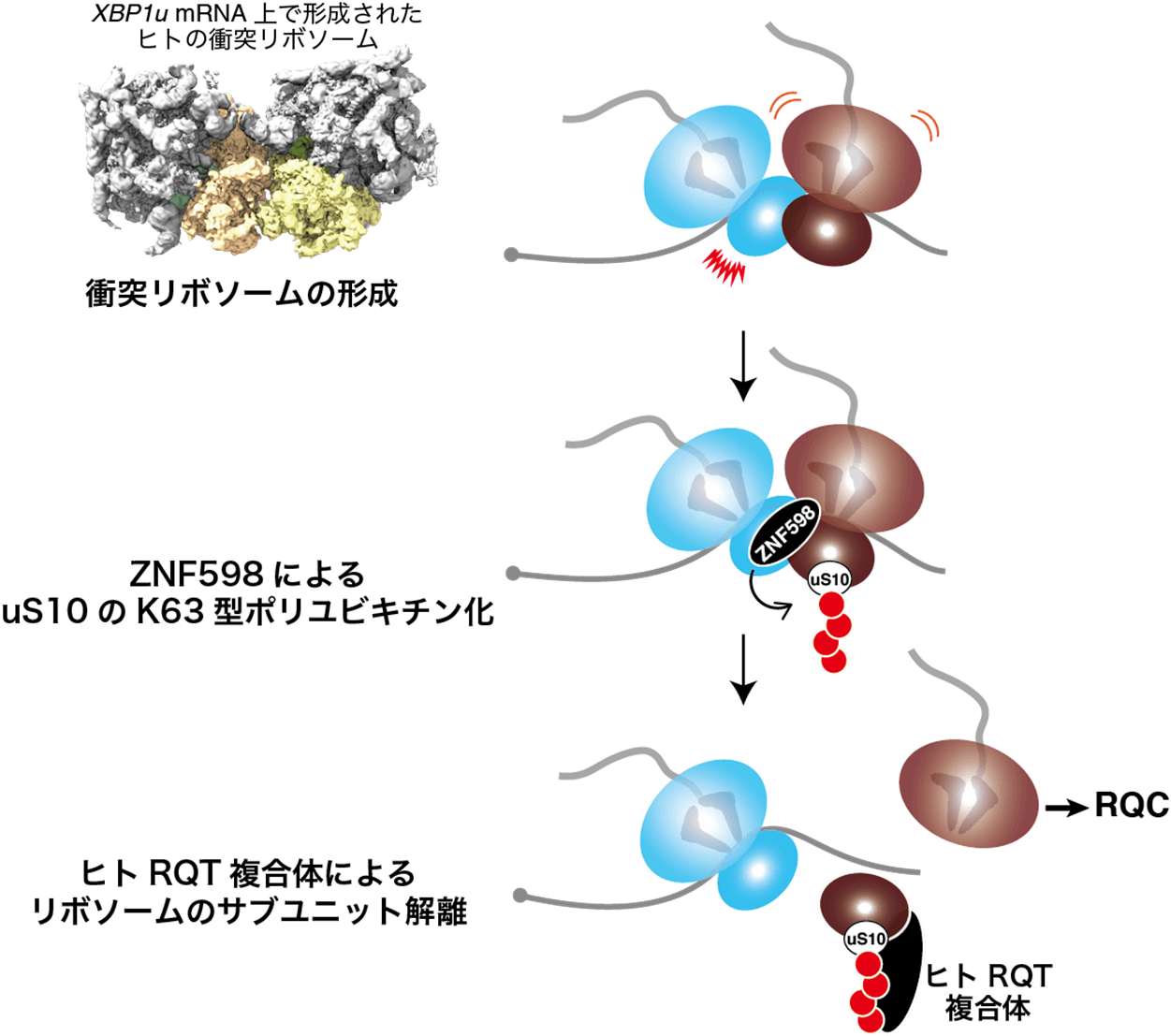
Using in vitro reconstitution experiments with endogenous translation stagnation sequences, the research group discovered the K63-type polyubiquitin modification as a novel ubiquitin modification of collision ribosomes in mammals, and revealed that the K63-type polyubiquitin modification is a landmark for induction of subunit dissociation of collision ribosomes The K63 polyubiquitin modification was found to induce subunit dissociation of collision ribosomes. Furthermore, the structure of the human collision ribosome was determined for the first time by structural analysis using cryo-electron microscopy. We also found differences from the collision ribosome structure of budding yeast, and discovered that the human collision ribosome may have acquired a different regulatory mechanism from that of budding yeast in evolution.
These findings are expected to lead to a better understanding of the pathogenic mechanisms of various neurological diseases associated with the accumulation of collision ribosomes and to the development of novel therapeutic strategies.
This research was supported by the Japan Agency for Medical Research and Development (AMED-CREST project number: 20gm1110010h0002, PI: Toshifumi Inada), JSPS Grants-in-Aid for Scientific Research (project numbers: 19H05281, 21H05277, 22H00401, Toshifumi Inada; 21H00267, 21H 05710, Yoshitaka Matsuo), and Japan Science and Technology Agency (JST) PRESTO (project number: JPMJPR21EE, PI: Yoshitaka Matsuo).
The research results have been published in the online edition of the U.S. scientific journal Nature Communications on October 27.
Figure: Molecular mechanism model of RQC in mammals
When collision ribosomes are formed in mammals, ZNF598 recognizes the collision and polyubiquitinates uS10 into K63-type polyubiquitin. The human RQT complex recognizes this K63-type polyubiquitin chain and performs subunit dissociation of the ribosome.
For more information, please visit the website of the Institute of Medical Science, The University of Tokyo.