DATE2022.09.23 #Press Releases
Make the cells glow with two-faced sensors!
Disclaimer: machine translated by DeepL which may contain errors.
- Development of a Chemical Genetics Sensor to Control Protein Fluorescence with Synthetic Molecules -.
Wenchao Zhu (Doctoral Student, Department of Chemistry)
Shiori Takeuchi (Master's course, Department of Chemistry)
Watayo Imai (4th year student at Department of Chemistry, Faculty of Science)
Takuya Terai ( Department of Chemistry, Project Associate Professor )
Robert Earl Campbell (Professor, Department of Chemistry / Professor, University of Alberta)
Key Points of the Presentation
- By appropriately combining small molecule chelators and fluorescent proteins, we have developed novel fluorescent sensors for calcium and sodium ions.
- Our unique design strategy, which combines synthetic organic chemistry and protein engineering, has enabled us to develop a sensor with properties that cannot be achieved by existing sensors using only proteins.
- With further improvement of performance and expansion of detection targets, this technology is expected to be useful in contributing to the elucidation of the mechanisms of brain functions and various diseases.
Summary of the presentation
Fluorescent biosensors that visualize the dynamic behavior of ions and molecules in vivo are one of the most important tools in modern biology. In this study, Graduate Student Wenchao Zhu, Project Associate Professor Takuya Terai, and Professor Robert Earl Campbell of The University of Tokyo Graduate School of Science have developed a new type of fluorescent sensor that combines green fluorescent protein (GFP) (Note 1) and a synthetic small molecule chelator (Note 2). This kind of two-fold design, so to speak, combining chemistry and biology is called chemogenetics. Although chemogenetic sensors themselves have been reported in the past, in this study, the part that binds to the target is made of a synthetic small molecule, which is thought to be more versatile than existing designs, allowing it to target a wider range of substances.
The HaloGFP-Ca1 developed in this study showed an 11.3-fold increase in fluorescence intensity upon binding of calcium ions (Ca) and was confirmed to actually function as a Ca sensor in cultured cells. Continued development of new sensors that take advantage of the sensor's improved performance and versatility is expected to contribute to the advancement of biological research, including the elucidation of brain and disease mechanisms.
Content of Presentation
Fluorescent biosensors are molecules that change their fluorescence intensity and color in response to changes in the concentration of specific substances in the body or the progress of biochemical reactions. For example, Ca is an important ion as a second messenger (Note 3) in the body, and many fluorescent biosensors have been developed to monitor changes in its concentration. They can be broadly classified into two types: sensors based on natural proteins and those using synthetic small molecules. The former has the advantage that it is genetically encoded and can be localized in cells and tissues, and its performance can be greatly enhanced by using a protein engineering technique called directed evolution, while the latter has the problem of being limited in the number of ions and molecules that can be targeted by the sensor. The latter is a problem because it is limited in the number of ions and molecules that can be targeted by the sensor. The latter is composed solely of artificial molecules produced by chemical synthesis, and while it has a wide range of detection targets, it is difficult to localize them in vivo. Recently, several chemigenetic (chemigenetic) sensors that combine proteins and synthetic molecules have been reported, but all of them are designed to bind small fluorescent molecules to some kind of protein, and there have been no chemigenetic sensors that use fluorescent proteins. The presenters reported that they have developed a sensor that can detect the target (chemigenetic).
In this study, the presenters devised and demonstrated a completely new chemical genetics sensor design in which the binding site to the target is a "small molecule" and the fluorescent site is a "fluorescent protein. The advantages of this design include the use of a protein, which allows for localization and directed evolution, and the fact that the target binding site is a small molecule, which allows for a wide range of detection targets and versatility.
The team first worked to develop a Ca2+ sensor that combines a synthetic small molecule chelator (BAPTA), which selectively binds Ca2+, with GFP. The chelator was attached to GFP using HaloTag, a protein tag that can incorporate specific small molecules (chloroalkanes). BAPTA is immobilized near the GFP chromophore by HaloTag, and the binding of Ca2+ to BAPTA is thought to alter the fluorescence intensity of GFP (Figure 1). To optimize the positional relationship between the chromophore and the chelator, we created 32 protein mutants in which cpGFP ( see(Note 1 )) was inserted into HaloTag. BAPTA was modified by adding a chloroalkane to allow it to bind to HaloTag. Ten variants were synthesized with different positions of the chelator and chloroalkane and different linker lengths, and the fluorescence intensity in the presence or absence of Ca2+ was measured in a total of 320 combinations of these variants and the 32 proteins described above. The combination that showed the largest change in fluorescence was 2.45-fold.
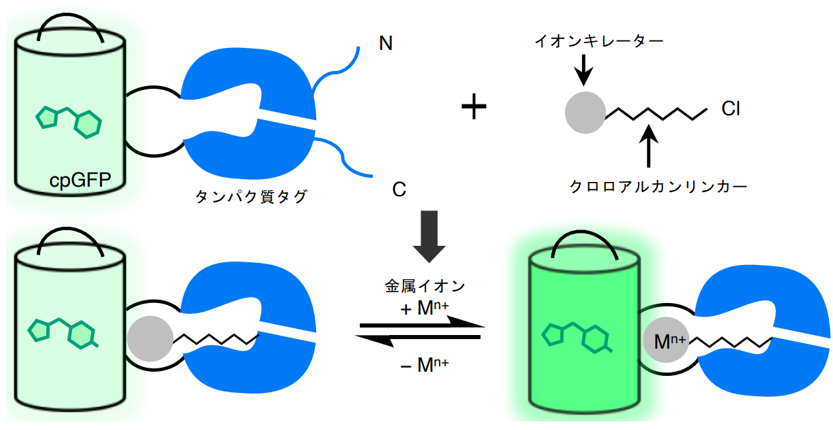
Figure 1: Schematic of the sensor design developed in this study. The binding of metal ions to the chelator of the synthetic small molecule changes the fluorescence intensity of GFP.
We then attempted to improve the performance using the directed evolution method, a protein engineering technique. By combining selective mutagenesis of specific residues with random mutagenesis of the entire gene, we finally achieved an 11.3-fold increase in fluorescence change and named this mutant HaloGFP-Ca1. HaloGFP-Ca1 was then expressed in HeLa cells (Note 4) and primary cultured neurons for Ca2+ imaging. In both cells, HaloGFP-Ca1 was confirmed to function as a Ca2+ sensor and was found to be capable of intracellular localization (Figure 2). To elucidate the mechanism of the sensor, simulations using molecular dynamics (Note 5) showed that the interaction between the carboxy group of BAPTA and residues on the protein is reduced when Ca2+ is bound, which may be one of the reasons for the fluorescence change (Figure 3).
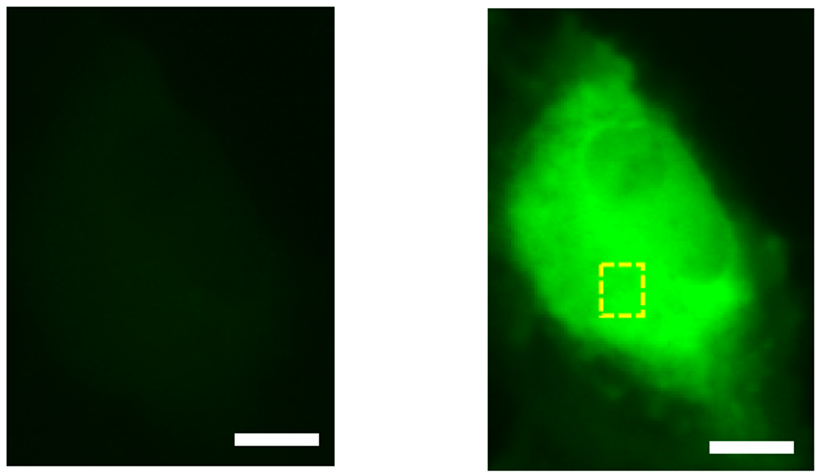
Figure 2: HeLa cell imaging with HaloGFP-Ca1. Compared to normal (left), fluorescence is stronger after addition of Ca2+ solution (right). Scale bar is 10 μm.
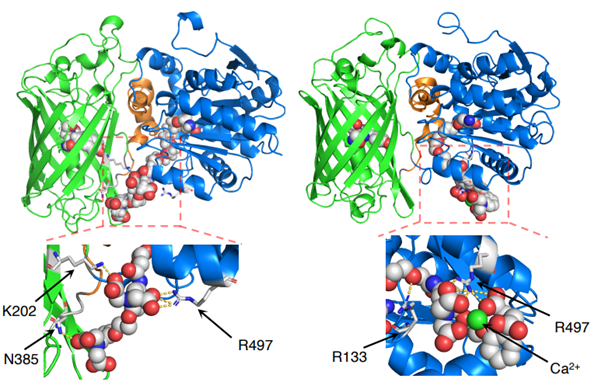
Figure 3: Simulation-predicted conformation of HaloGFP-Ca1, which is expected to show the conformational changes shown in the figure before (left) and after (right) Ca2+ binding.
Although HaloGFP-Ca1 has a large change in fluorescence, it has a problem in that it has a low affinity for Ca2+. The research team addressed that issue as well by using directed evolution to develop a new mutant, HaloGFP-Ca0.2, which responds adequately at physiological Ca2+ concentrations in the cell. To demonstrate the versatility that is an advantage of this design, we also developed a sensor for another metal ion that has important functions in vivo, sodium ion ( Na+). cpGFP was inserted into HaloTag using the same procedure as HaloGFP-Ca1 and instead of BAPTA, a Na+ As with HaloGFP-Ca1, we evaluated each protein + ligand combination and then used directed evolution to improve the performance, and succeeded in developing HaloGFP-Na0.5 which shows ΔR/R0 (Note 6) = 286% by Na+ binding, indicating that the sensor design method can be applied to more than just Ca2+.
Future work includes the development of sensors that target other ions and biomolecules, as well as sensors using red fluorescent protein (RFP), which has a wavelength more suitable for cellular experiments. Continued enhancement of these targets and performance improvements is expected to lead to the creation of many new fluorescent sensors that will replace conventional fluorescent sensors. Fluorescent sensors are now indispensable tools for visualizing the complex reactions that occur in cells. The emergence of new sensors that overcome the shortcomings of conventional sensors will further advance research in biology, where much is still unknown, such as the mechanisms of the brain and the mechanisms of disease.
This work was supported by Grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (19H05633, Robert E. Campbell; 18H02103, 21H00273, Takuya Terai).
The main members of this study:
| Wenchao Zhu | D. Department of Chemistry, Graduate School of Science, The University of Tokyo |
| Shiori Takeuchi | M.S. Department of Chemistry, Graduate School of Science, The University of Tokyo |
| Watayo Imai | The University of Tokyo, Faculty of Science, Department of Chemistry, 4th year |
| Toru Terada | Associate Professor, Department of Biotechnology, Graduate School of Agricultural and Life Sciences, The University of Tokyo |
| Takumi Ueda | Associate Professor, Department of Pharmacy Sciences, Graduate School of Pharmaceutical Sciences, The University of Tokyo |
| Yusuke Nasu | Assistant Professor, Department of Chemistry, Graduate School of Science, The University of Tokyo |
| Takuya Terai | Project Associate Professor, Department of Chemistry, Graduate School of Science, The University of Tokyo |
| Robert Earl Campbell | (Professor, Department of Chemistry, Graduate School of Science, The University of Tokyo / Professor, Department of Chemistry, The University of Alberta) |
Journals
-
Journal name Nature Chemical Biology Title of paper Chemigenetic indicators based on synthetic chelators and green fluorescent protein Author(s) Wenchao Zhu , Shiori Takeuchi , Shosei Imai , Tohru Terada , Takumi Ueda , Yusuke Nasu , Takuya Terai* & Robert Campbell* (author) DOI Number
Terminology
1 Green fluorescent protein (GFP)
A protein that exhibits green fluorescence, isolated in 1962 by Dr. Osamu Shimomura (Nobel Prize in Chemistry 2008) from a jellyfish. The part of the protein that emits fluorescence is called the chromophore. In this study, cpGFP (circularly permuted green fluorescent protein), whose terminal position was changed near the chromophore of GFP to facilitate fluorescence change, was used. ↑up
Note 2: Chelator
A molecule that can bind to specific metal ions. It has multiple metal-binding sites, which enable it to strongly capture metal ions. BAPTA used in this study is known as a chelator that can selectively bind Ca2+ with four carboxy groups, two amino groups, and two oxygen atoms. Also, crown ether is a molecule consisting of a carbon atom and an oxygen atom connected in a ring, and depending on the size of the ring, it can take in various ions such as Na+. ↑up
Note 3 Second messenger
In the transmission of information in living organisms, the exchange of information between cells is carried out by molecules called first messengers, such as hormones. However, many of these molecules cannot pass through the cell membrane, so intracellular signaling is carried out by another group of molecules called second messengers. ↑up
Note 4 HeLa cells
Cells isolated from human cervical cancer in 1951. Because of their extremely high proliferative potential, HeLa cells are cultured worldwide and used in numerous studies. ↑up
Note 4 Molecular dynamics
A simulation method that tracks the overall motion of all the particles (atoms) that make up an object from moment to moment according to the equations of motion by repeating calculations of what forces they are subjected to from their surroundings. ↑up
Note 4 ΔR/R0
GFP can fluoresce with two types of light, light with a wavelength of approximately 400 nm and light with a wavelength of 500 nm; R is the ratio of the intensity of fluorescence from 400 nm light to that from 500 nm light, and ΔR/R0 indicates the percentage change in the value of R before and after target binding. ↑↑


