DATE2022.06.24 #Press Releases
Developed a method to estimate mechanical parameters of epithelial cells from image data
Disclaimer: machine translated by DeepL which may contain errors.
Takeshi Ogita (Technical Assistant, Department of Biological Sciences)
Takeshi Kondo, Lecturer, Graduate School of Life Sciences, Kyoto University
Keisuke Igawa, Project Assistant Professor, Department of Biological Sciences
Tadashi Kamimura, Professor, Graduate School of Life Sciences, Kyoto University
Hideyuki Ishihara, Associate Professor, Graduate School of Arts and Sciences, The University of Tokyo
Kaoru Sugimura, Associate Professor, Department of Biological Sciences
Key points of the presentation
- We have developed a fast and simple method for estimating mechanical parameters representing mechanical properties (Note1 ) of epithelial cells (Note2) from images (Figure 1).
- In this study, we applied the developed method to Drosophila epithelial tissue and found that a mechanical parameter, the negative spring constant of the cell adhesion surface, can promote cell reordering (Figs. 2 and 3).
- Because of its simplicity, the developed method is expected to be applied to gene screening (Note 3) based on a new index of mechanical parameters.
Summary of the presentation
The body of a living organism is formed by repeated, orderly deformation of tissues. In this process, cells are known to regulate the speed and direction of deformation by changing mechanical properties such as elasticity and viscosity, in addition to exerting forces themselves to drive the deformation. Advances in experimental techniques over the past decade or so have made it possible to measure deformation and force in cells and tissues, even in living individuals. On the other hand, in vivo measurement of mechanical properties of cells has required advanced techniques.
In this study, a research group led by Technician Goshi Ogita and Associate Professor Kaoru Sugimura of the Department of Biological Sciences, Graduate School of Science, The University of Tokyo, has developed a fast and simple method to estimate mechanical parameters representing mechanical properties of epithelial cells from image data. Tests using artificial data generated by simulation have confirmed that the method developed in this study can estimate the parameters more accurately than conventional methods. Furthermore, by applying the developed method to Drosophila epithelial tissue, it was shown that a mechanical parameter, the negative spring constant of cell adhesion surfaces, can promote cell rearrangement. The simplicity of this method makes it applicable to large-scale genetic screening, and it is expected to become a powerful tool in mechanobiology (Note 4).
Contents of the presentation
Background of the research and problems in previous studies
The body of an organism is formed by repeated, orderly deformation of tissues. In this process, cells are known to regulate the speed and direction of deformation by changing mechanical properties such as elasticity and viscosity, in addition to exerting forces themselves to drive the deformation. Advances in experimental techniques over the past decade or so have made it possible to measure deformation and force in cells and tissues, even in living individuals. In contrast, in vivo measurements of the mechanical properties of cells have required sophisticated techniques.
As an alternative to experimental measurements, research using theoretical models has been conducted in parallel. In the approach using theoretical models, a model representing the mechanical properties of cells is constructed, simulations are performed, and the obtained tissue morphology is compared with the actual tissue morphology to investigate the relationship between the mechanical properties of cells and tissue morphology.
On the other hand, there were several problems in the construction of models and parameter estimation in previous studies. Specifically, the model functions are determined based on qualitative knowledge of molecular functions, and the parameters are estimated by indirectly comparing the simulation results with actual tissues via summary statistics such as the polygonal distribution of cells. Against this background, this study aimed to develop a method to construct models based on quantitative data and to estimate mechanical parameters directly from the information possessed by the data.
Description of Research
In this study, we developed a fast and simple method for estimating mechanical parameters representing mechanical properties of epithelial cells from image data (Figure 1). The developed method consists of model building, parameter estimation, and model evaluation, and outputs the optimal mechanical model and mechanical parameters of epithelial tissue based on the input image data of fluorescently labeled cell adhesion surfaces of epithelial tissue. In model building, the functional form of the mechanistic model is determined based on the quantitative correlation between cell forces and morphological features estimated from the image data by the Bayesian estimation method of force (Note 5) (Figure 2). Next, the parameters of the model function are estimated by substituting the model function into the force balance equation at the cell apex and solving for the parameters. Finally, the Akaike Information Criterion, a model evaluation criterion widely used in statistics, is used to select the most appropriate mechanics model among multiple model functions.
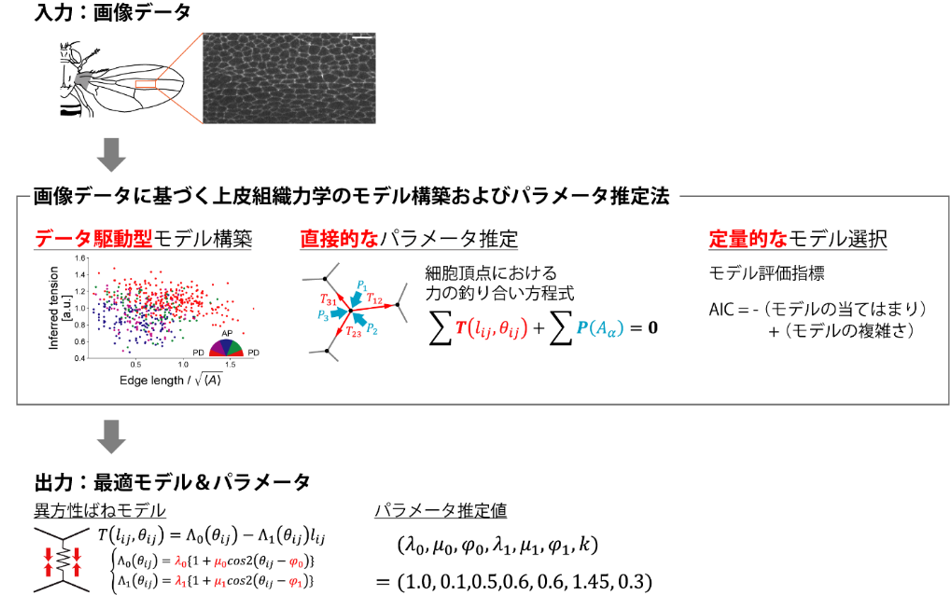
Figure 1: Overview of the method developed in this study
The method takes image data (upper panel) as input and outputs the best-fit mechanics model and mechanics parameters (lower panel). The method first constructs a function of the mechanics model from the relationship between the force and morphological features of the cell estimated from the image data by the Bayesian estimation method of force (middle row, left; Figure 2 for details). Next, the constructed function of the mechanical model is substituted into the equation of force balance at the cell apex, and the estimated mechanical parameters are obtained by solving this equation (middle row, center). Finally, the most suitable mechanics model is selected among several mechanics models by the minimum AIC method (middle row, right).
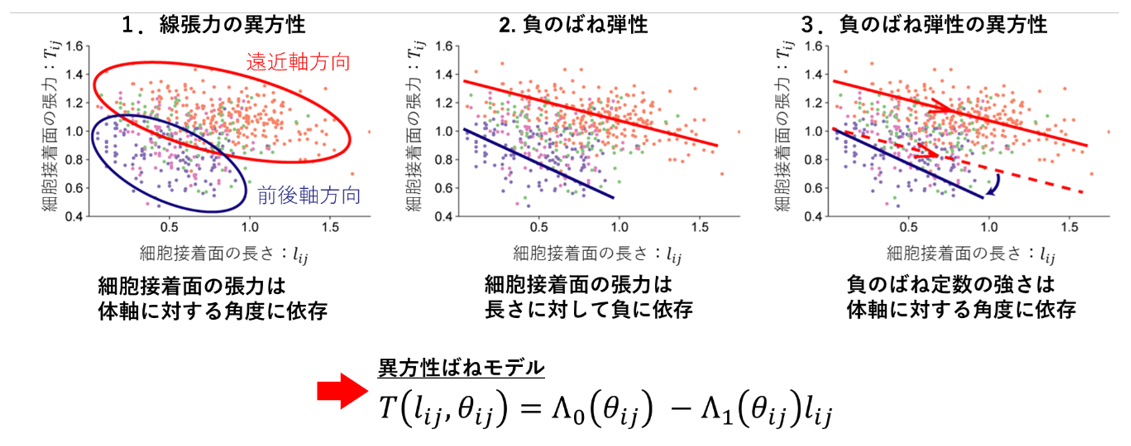
Figure 2: Mechanical model of cell adhesion surface constructed by the method developed in this study.
The function system of the mechanical model was determined based on the characteristics of the relationship between the tension, length, and orientation of the cell adhesion surface estimated by the Bayesian force estimation method. The upper graph shows the relationship between the tension (vertical axis), length (horizontal axis), and orientation (color of the dots) of the cell adhesion surface. A mechanical model of the cell adhesion surface (anisotropic spring model) was constructed based on the data by expressing the features shown in the upper graph in the lower equation.
The mechanical model of cell adhesion surfaces (anisotropic spring model) constructed in this study has the following features. First, the magnitude of the tension of the cell adhesion surface depends on the angle of the cell adhesion surface (anisotropy of linear tension). Second, the tension of the cell adhesion surface shows a negative dependence on the length of the cell adhesion surface (negative spring elasticity). Third, the negative spring elasticity of the cell adhesion surface depends on the angle of the cell adhesion surface (anisotropy of negative spring elasticity).
Tests with artificial data generated using simulations show that accurate parameter estimation and appropriate model selection are possible using this method. It was also confirmed that the estimation using this method is robust against errors in image analysis.
Next, the developed method was applied to Drosophila epithelial tissue to analyze the mechanical properties of Drosophila epithelial cells. Using the Akaike information criterion, the mechanical model commonly used in the field was compared with the anisotropic spring model, and the latter was selected for all samples, suggesting that the anisotropic spring model better represents the mechanical properties of Drosophila epithelial tissue. The estimation of mechanical parameters using the anisotropic spring model revealed that the mechanical parameters change with the developmental stage of Drosophila. Furthermore, by comparing the estimated mechanical parameters with what is known about epithelial tissue mechanics and morphogenesis, it was clarified that the angular dependence of the linear tension of the cell adhesion surface plays a role in resisting forces from outside the tissue, while the angular dependence of the negative spring constant of the cell adhesion surface plays a role in promoting cell rearrangement (Figure 3).
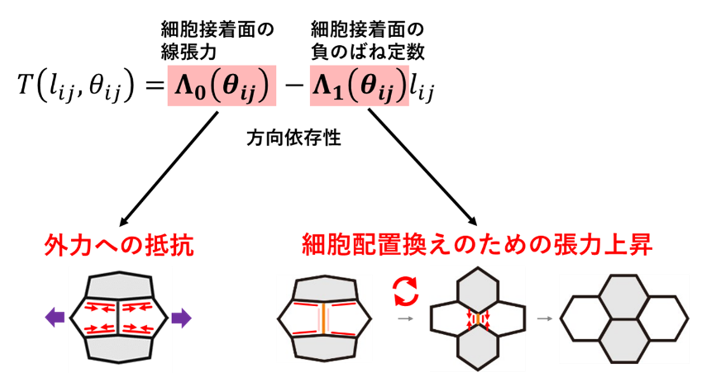
Figure 3: Role of anisotropy of mechanical properties of cell adhesion surfaces suggested by analysis of Drosophila epithelium.
Comparison of the estimated mechanical parameters with what is known about epithelial tissue mechanics and morphogenesis reveals that the angular dependence of the linear tension of the cell adhesion surface plays a role in resisting forces from outside the tissue, while the angular dependence of the negative spring constant of the cell adhesion surface plays a role in promoting cell reordering.
Future Developments
Unlike the methods used in previous studies, the method developed in this study builds a model based on quantitative information in the form of correlation between cell forces and morphological features, and estimates parameters directly from the data. Furthermore, since the parameter estimation in this method does not require repeated simulations, the mechanical parameters can be estimated quickly and easily from image data. Therefore, by applying this method to various tissues of various organisms in the future, it may become a useful tool for elucidating how epithelial tissues form various shapes and the principles common to the morphogenesis of epithelial tissues. Furthermore, it is also expected to be applied to genetic screening to search for genes involved in the mechanical control of morphogenesis using the estimated mechanical parameters as indicators.
This research was supported by
JSPS Grant-in-Aid for Scientific Research (17K15125), JSPS Research Grant (18H01185), JSPS International Collaborative Research Program with Switzerland (JPJSJPR20191501), and Innovative Advanced Research and Development Support from National Institute of Biomedical Innovation Project (20gm5810025h9904), Japan Science and Technology Agency's Strategic Creative Research Promotion Program (JPMJCR1923)
Journals
-
Journal name PLOS Computational Biology Title of paper Image-based parameter inference for epithelial mechanics Author(s) Goshi Ogita, Takefumi Kondo, Keisuke Ikawa, Tadashi Uemura, Shuji Ishihara*, Kaoru Sugimura*, and Shuji Ishihara DOI number
Terminology
1 Epithelial cells
Cells that make up epithelial tissue. Epithelial tissue covers the surface of the body of living organisms and acts as a barrier between the outside world and the inside of the body. An example is the intestinal epithelium in humans. To achieve the barrier function, cells are tightly adhered to each other via intercellular adhesion molecules. During the development of an individual, epithelial cells remain adherent to each other and dynamically change their arrangement to promote tissue deformation and multicellular pattern formation. ↑up
Note 2 Mechanical properties
A physical property of a material that determines how much it deforms when a force is applied and whether or not it returns to its original shape when the force is removed. Typical examples include elasticity and viscosity. ↑up
Note 3 Gene screening
An experimental method to identify genes involved in a certain biological phenomenon. In sequential genetic screening, individuals that show abnormalities in the life phenomenon of interest are isolated from a population of organisms whose genomes have been randomly mutated. Then, by identifying the mutations in the isolated individuals, the genes involved in the biological phenomenon of interest are identified. ↑up
Note 4 Mechanobiology
A field of study that elucidates the role played by the dynamics of molecules, cells, and tissues in biological phenomena. ↑↑
Note 5 Bayesian Estimation of Forces
A method previously proposed by this research group to estimate the relative values of tension on cell adhesion surfaces and pressure on cells from images of epithelial tissues (Ishihara and Sugimura. Journal of Theoretical Biology 313, 201-211, 2012; https:// doi.org/10.1016/j.jtbi.2012.08.017). ↑


