DATE2022.06.30 #Press Releases
How the Dicer-2-R2D2 protein complex senses double-stranded siRNA asymmetry.
Disclaimer: machine translated by DeepL which may contain errors.
- Solving a 20-Year-Old Mystery in RNA Interference - The
Sonomi Yamaguchi (Department of Biological Sciences, 3rd year of Doctoral program)
Yukihide Tomari (Professor, Institute for Quantitative Biosciences)
Hiroshi Nishimasu (Professor, Research Center for Advanced Science and Technology)
Osamu Nureki (Professor, Department of Biological Sciences)
Key points of the presentation
- The steric structure of the complex formed from Drosophila Dicer-2, its partner protein R2D2, and double-stranded siRNA was clarified for the first time in the world.
- The molecular mechanism by which Dicer-2-R2D2 passes double-stranded siRNA to Argonaute in a fixed orientation has been clarified.
- This achievement is expected not only for understanding the working mechanism of Dicer, but also for technological applications in the design of efficient double-stranded siRNA and nucleic acid medicine.
Summary of Presentation
A small RNA of about 20 bases (siRNA (Note 1) ) forms an effector complex with Argonaute protein and regulates the expression of a target gene with a complementary nucleotide sequence. This gene regulation mechanism is called RNA interference, which acts as an immune mechanism in insects such as Drosophila. Furthermore, RNA interference is also widely used as a technology to suppress gene expression, and the RNA-cleaving enzyme Dicer-2 is responsible for the production of double-stranded siRNA and its delivery to Argonaute. However, its detailed molecular mechanism has not been clarified.
In this study, Somi Yamaguchi, a graduate student, Professor Osamu Nureki of the Department of Integrated Biosciences, Professor Yukihide Tomari of the Institute for Quantitative Biosciences, Professor Hiroshi Nishimasu of the Research Center for Advanced Science and Technology, and their colleagues at the Graduate School of Biological Sciences, The University of Tokyo, used a cryo-EM (Note 2) to determine the steric structure of the Dicer-2-R2D2 double-stranded siRNA complex using cryo-electron microscopy (Note 2). As a result, the molecular mechanism of double-stranded siRNA production and delivery by Dicer-2-R2D2 was clarified. This achievement is expected to be applied not only to the understanding of the working mechanism of Dicer, but also to the design of efficient double-stranded siRNAs and nucleic acid medicine (*3) and other technologies.
Announcement
Drosophila Argonaute (Ago2) forms an effector complex called RISC (RNA-induced silencing complex) with siRNA to repress expression of RNA from viruses and transposons (Note 4) (Figure 1). Dicer-2 forms a complex with its partner protein R2D2 to cleave long double-stranded RNA to produce double-stranded siRNA (Figure 1). The thermodynamic stability of the ends of double-stranded siRNA differs depending on the base sequence. In other words, the ends that contain many mismatched bases or A-T base pairs are thermodynamically unstable. Biochemical analysis by Professor Tomari and his colleagues in 2004 revealed that Dicer-2-R2D2 not only cleaves long double-stranded siRNA, but also senses the asymmetry of the produced double-stranded siRNA once it is released and recombines it so that the thermodynamically unstable end is near Dicer-2 and the stable end is near R2D2. (Fig. 1). Furthermore, biochemical analysis showed that Ago2 selects the RNA strand containing the thermodynamically unstable end of the double-stranded siRNA as the guide strand (Note 5) (Figure 1).
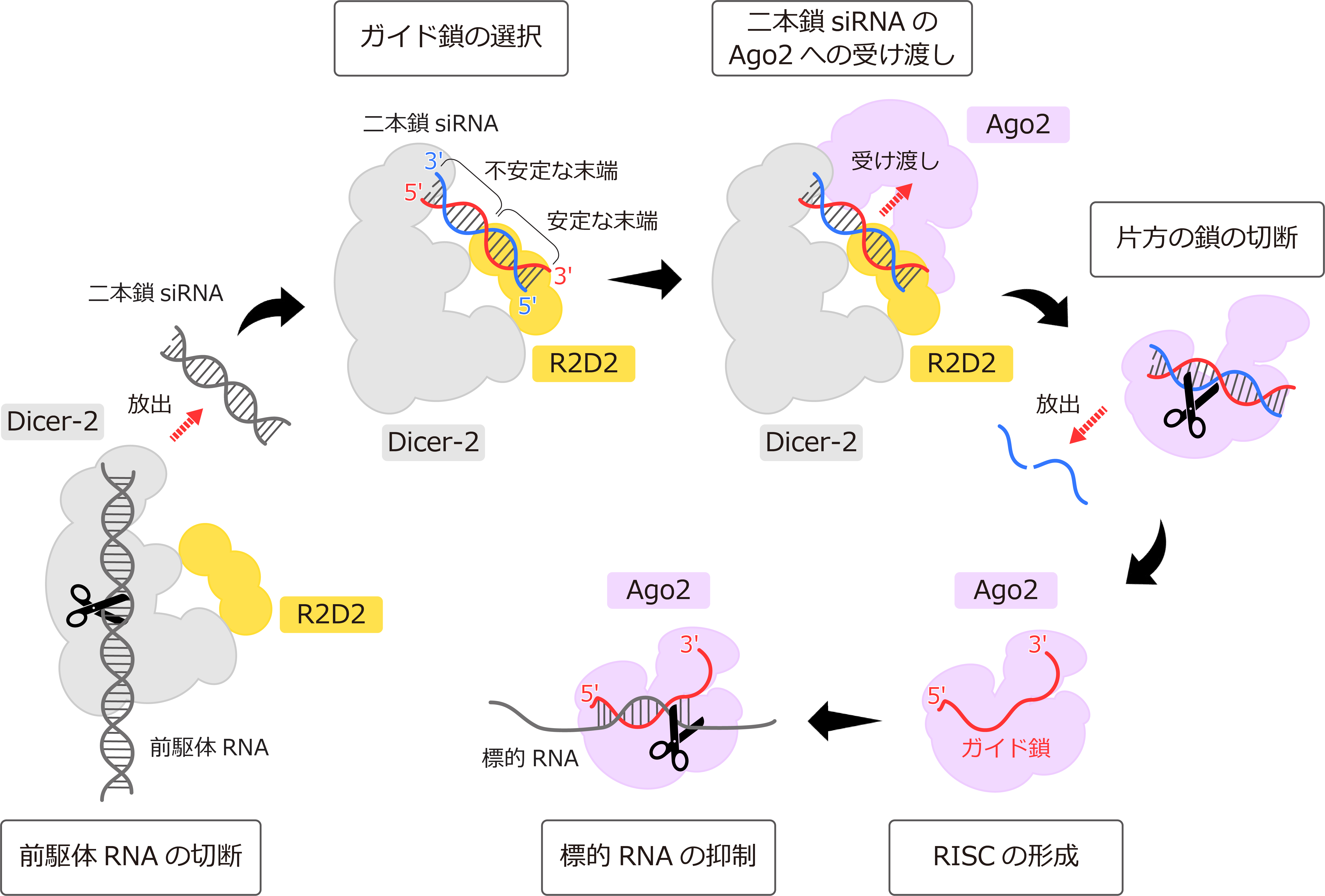
Figure 1: Function of Dicer-2-R2D2 and Ago2 in RNA interference.
Therefore, it has been suggested that Dicer-2-R2D2 senses the thermodynamic asymmetry of double-stranded siRNA and determines which strand of double-stranded siRNA is used as the guide strand by passing the double-stranded siRNA to Ago2 in a specific orientation. Since the sequence of the guide strand determines which gene is repressed in vivo, delivery of double-stranded siRNA to Ago2 by Dicer-2-R2D2 is an important step in determining the target gene. However, the steric structure of Dicer-2-R2D2 is unknown, and the molecular mechanism by which Dicer-2-R2D2 senses the thermodynamic asymmetry of double-stranded siRNA and delivers double-stranded siRNA to Ago2 in a specific orientation has not been clarified for nearly 20 years since the discovery of R2D2 The molecular mechanism that senses thermodynamic asymmetry and delivers double-stranded siRNA to Ago2 in a fixed orientation has not been clarified for nearly 20 years since the discovery of R2D2.
In this study, we determined the steric structure of double-stranded siRNA bound to the Dicer-2-R2D2 protein complex at 3.3 Å resolution using cryo-EM. The results revealed that Dicer-2 is mainly composed of a helicase domain, Platform-PAZ domain, RNase III domain, and C-terminal RNA-binding domain (CRBD), and that it binds to R2D2 via the helicase domain. Notably, double-stranded siRNA in a guided strand selection state bound to R2D2 (Figure 2).
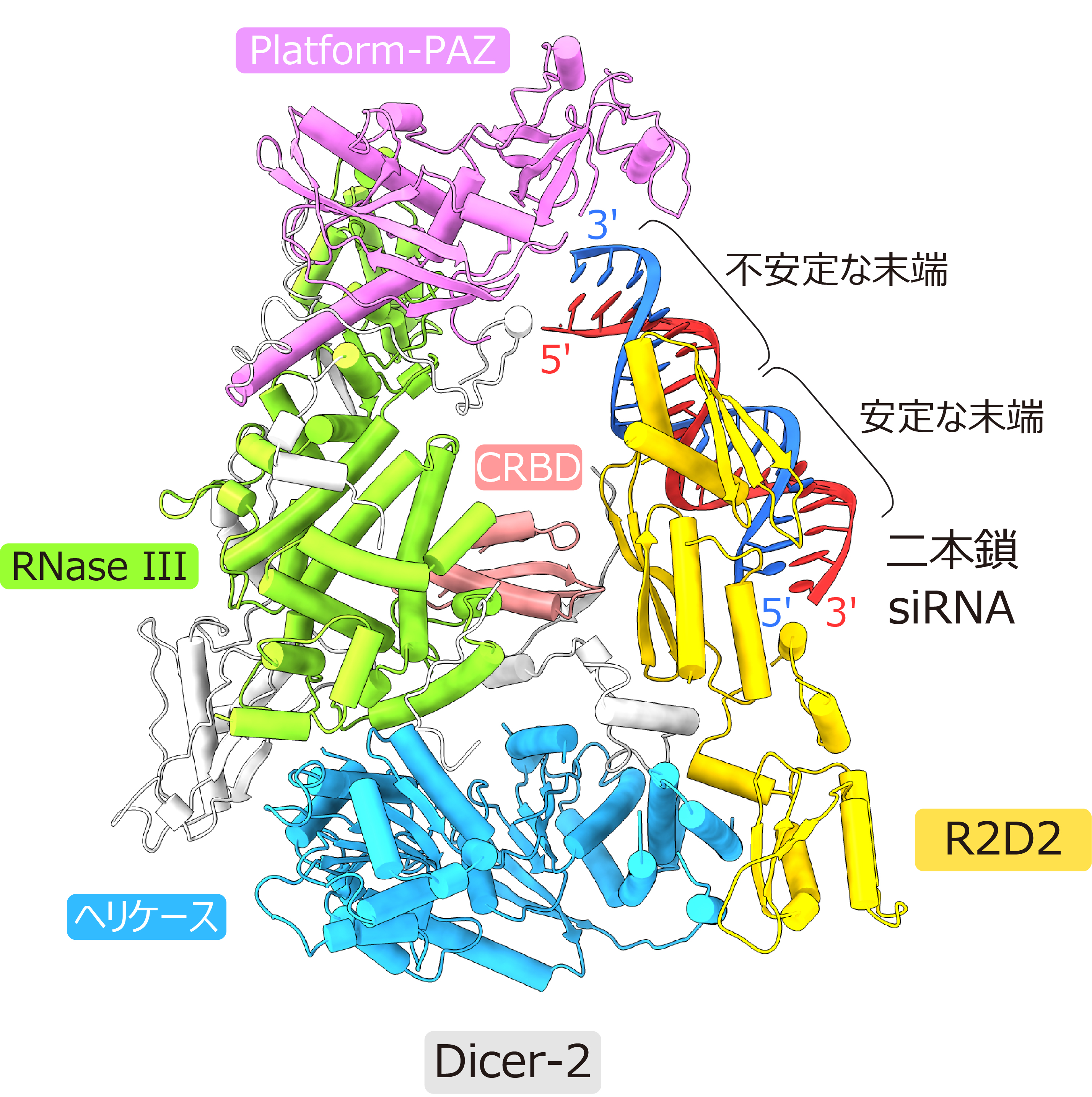
Figure 2: Cryo-EM structure of the Dicer-2-R2D2-double-stranded siRNA complex.
The thermodynamically unstable end of the double-stranded siRNA was exposed to the solvent, while the stable end was bound to R2D2. Biochemical analysis revealed that R2D2 binds preferentially to the more double-helical (i.e., thermodynamically stable) end. A model of Ago2 binding to the present structure was created, suggesting that the unstable end of the double-stranded siRNA is in a suitable position for binding to Ago2 (Figure 3).
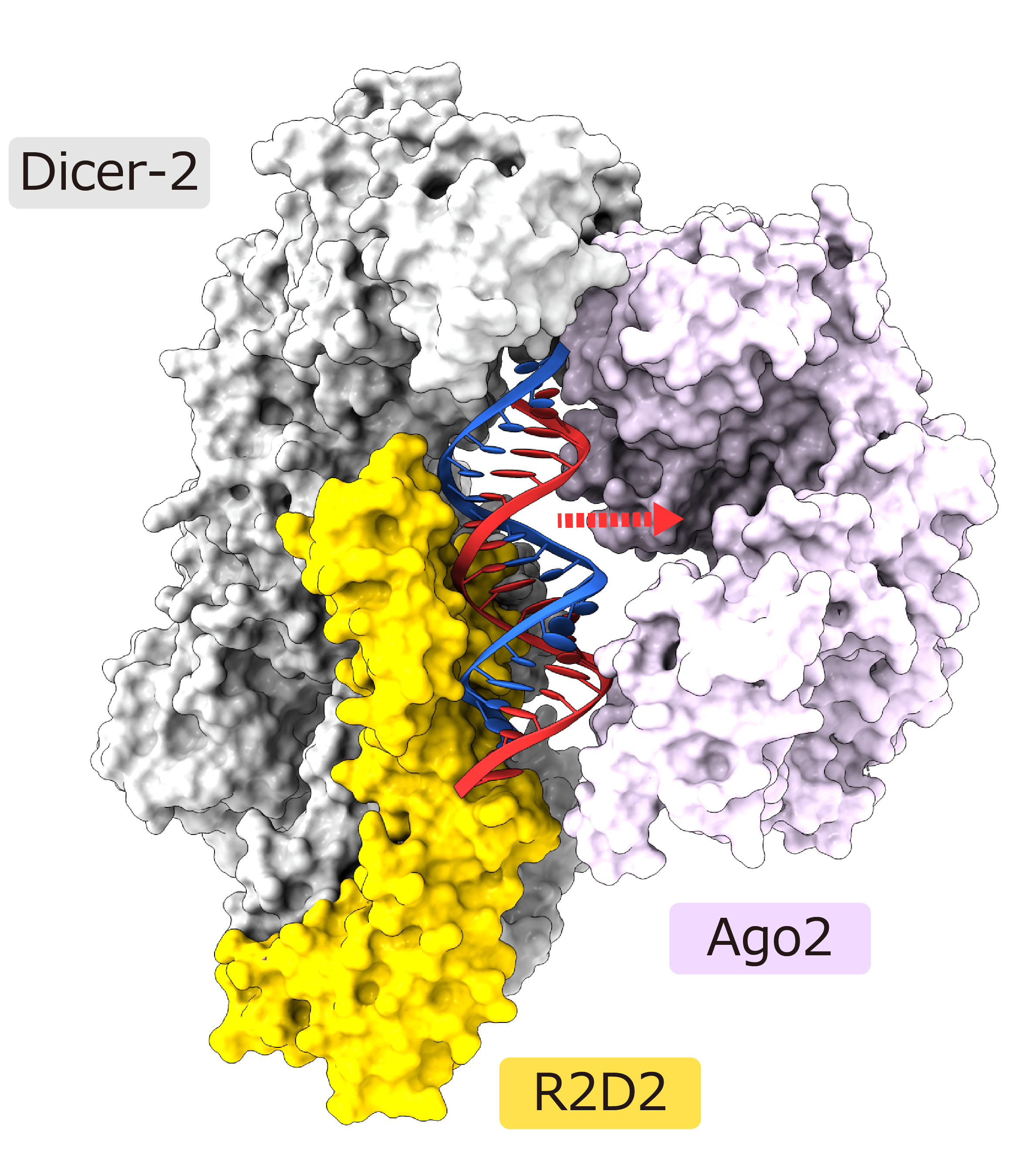
Figure 3: Structural model of Ago2 bound to Dicer-2-R2D2-double-stranded siRNA complex
These results reveal the molecular mechanism by which Dicer-2-R2D2 delivers double-stranded siRNA to Ago2 in a specific orientation. The results of this study are expected to be applied to nucleic acid medicine.
This research was supported by CREST "Dynamics of Supramolecular Complexes for Cellular Functions" (Project Leader: Osamu Nureki, JPMJCR20E2), the Public-Private R&D Investment and Scaling Up Program (PRISM) "Construction of Functional Module Data Base for Genome Editing Enzymes" (Project Leader: Osamu Nureki, JPJ008000), the (PI: Osamu Nureki), JSPS Grants-in-Aid for Scientific Research (S) "Biochemical Analysis of piRNA Pathways Focusing on Reaction Fields" (Project number: 18H05271 PI: Yukihide Tomari), Fundamental Research (B) "Structure-function analysis of RNA-dependent effector complexes" (Project number: 18H02384 PI: Hiroshi Nishimasu), Special Research (Principal Investigator: Hiroshi Nishimasu), "Structure-function analysis of RNA silencing-related factors in Drosophila" (Research Fellowship: 21J11067, Special Researcher: Sonomi Yamaguchi), ANRI Basic Science Scholarship "Structure-function analysis of RNA silencing-related factors in Drosophila" (Sonomi Yamaguchi), etc. The research was supported by ANRI Basic Science Scholarship.
Journals
-
Journal name Nature Title of paper Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex Author(s) Sonomi Yamaguchi, Masahiro Naganuma, Tomohiro Nishizawa, Tsukasa Kusakizako, Yukihide Tomari, Hiroshi Nishimasu, Osamu Nureki DOI Number
Terminology
Note 1. siRNA
Abbreviation for small interfering RNA (siRNA), a type of small RNA. 21-23 bases long single-stranded RNA. up
2 Cryo-electron microscope
A device for observing samples by irradiating electron beams to molecules such as proteins under liquid nitrogen (-196°C) cooling. It is used to determine the three-dimensional structure of proteins and nucleic acids. ↑up
Note 3 Nucleic acid medicine
A generic term for pharmaceuticals that use nucleotides, the building blocks of DNA and RNA, as their basic backbone. They are expected to be next-generation drugs because they target nucleic acid molecules that cannot be targeted by conventional low-molecular-weight drugs or antibody drugs. ↑up
Note 4 Transposon
A gene that moves on the genome. Transposon transfer has the potential to lead to the destruction of genetic information. ↑up
Note 5 Guide strand
A strand of double-stranded siRNA that is complementary to the target RNA. Argonaute in which this strand is incorporated recognizes and cleaves the target RNA. ↑up


