DATE2022.04.25 #Press Releases
Molecular interaction networks that promote cell proliferation under insulin stimulation are elucidated from large scale measurement data of
biomolecules.
Disclaimer: machine translated by DeepL which may contain errors.
Ei Terakawa, 3rd Year Doctoral Student, Department of Biological Sciences
Yanhui Hu, Ph.D., Harvard Medical School
Norbert Perrimon (Professor, Harvard Medical School)
Shinya Kuroda, Professor, Department of Biological Sciences
Key Points of Presentation
- By integrating the molecular interaction network constructed from the multi-omics data of insulin-stimulated Drosophila cells (Note 1) with data on cell proliferation during comprehensive single gene knockout, we have identified a network involved in the regulation of proliferation under insulin stimulation.
- The data analysis method developed in this study enables large-scale evaluation of the relationship between molecular interaction networks and phenotype, which has been largely unexplored in previous studies.
- It is expected that the developed method will be used to elucidate the mechanisms of action (Note 2) of various life phenomena and diseases in the future.
Summary of the presentation
Cellular phenotypes such as proliferation and growth are regulated by large-scale molecular interaction networks including transcripts, proteins, and metabolites. With advances in omics measurement technology (*3) and data analysis methods, it has become possible to identify comprehensive intracellular molecular interaction networks. However, there has been no method to link such networks to phenotypes, which has been a barrier to elucidating the physiological significance of the identified networks and to considering their linkage to diseases.
D. students Ei Terakawa and Professor Shinya Kuroda at the Graduate School of Science, The University of Tokyo, Dr. Yanhui Hu and Professor Norbert Perrimon at Harvard Medical School, Assistant Professor Toshiya Kogajiri at the Nara Institute of Science and Technology's Data-driven Science Creation Center, and Team Leader Katsuyuki Yuzuki at the RIKEN Graduate School of Science's Center for Biomedical Sciences. In collaboration with Assistant Professor Toshiya Kogajiri at Nara Institute of Science and Technology, Team Leader Katsuyuki Yuzuki at RIKEN Biomedical Science Center, Associate Professor John M. Asara and Professor Martha L. Bulyk at Harvard Medical School, a large scale molecular interaction network (trans-omics network (*4) ) constructed from the integrated analysis of multi-omics data, and and CRISPR knockout screening data (Note 5) ( hereafter, CRISPR screening data), which are phenotypic measurement data at the time of gene knockout, we developed an integrated analysis method. In particular, in this study, we applied the developed method to insulin-stimulated multi-omics data obtained from Drosophila cells and CRISPR screening data of cell proliferation to clarify the trans-omics network involved in the regulation of cell proliferation under insulin stimulation.
The findings obtained in this study may lead to a better understanding of the pathogenesis and treatment of insulin-related diseases in the future. Furthermore, it is expected that the developed methods will be used to elucidate the mechanisms of action of various other life phenomena and diseases.
Contents of the presentation
Background of the research and problems in previous studies
Cellular phenotypes such as proliferation and growth are appropriately regulated by changing a large trans-omics network including transcripts, proteins, and metabolites in response to external environmental conditions and stimuli such as nutritional conditions and growth factors. To date, the development of omics measurement techniques has made it possible to comprehensively measure the abundance and interactions of these molecules under a variety of experimental conditions. In line with this development, methods have been actively developed to estimate trans-omics networks that are activated or inactivated under experimental conditions from the measured data. However, which subnetworks of these networks are involved in which phenotypes has not been evaluated, and one of the challenges in this field has been to link the obtained networks to the phenotypes.
Insulin is an important hormone that is evolutionarily highly conserved from Drosophila to mammals and has effects of lowering blood glucose levels and promoting cell proliferation and growth. Although a large transomics network regulated by insulin has been identified so far, including previous studies by this research group, the contribution of most of the network to phenotypes, including cell proliferation, has not yet been elucidated.
To address this issue, this research group has developed a method to integrate CRISPR screening data, which is a comprehensive measurement of cellular phenotypes upon single gene knockout, with the transomics network constructed from multi-omics data analysis, and has attempted to solve the above problem. In particular, in this study, we applied the developed method to insulin-stimulated multi-omics data and CRISPR screening data of cell proliferation in Drosophila cells, and clarified the trans-omics network involved in the regulation of cell proliferation under insulin stimulation.
Research Details
This research group 1) constructed a transomics network regulated by insulin using insulin-stimulated multi-omics data, and 2) performed an integrated analysis of the obtained transomics network and CRISPR screening data for cell proliferation to identify the transomics network involved in the regulation of cell proliferation under insulin stimulation. The transomics network involved in the regulation of cell proliferation under insulin stimulation was clarified by integrating the obtained transomics network with CRISPR screening data (Figure 1).
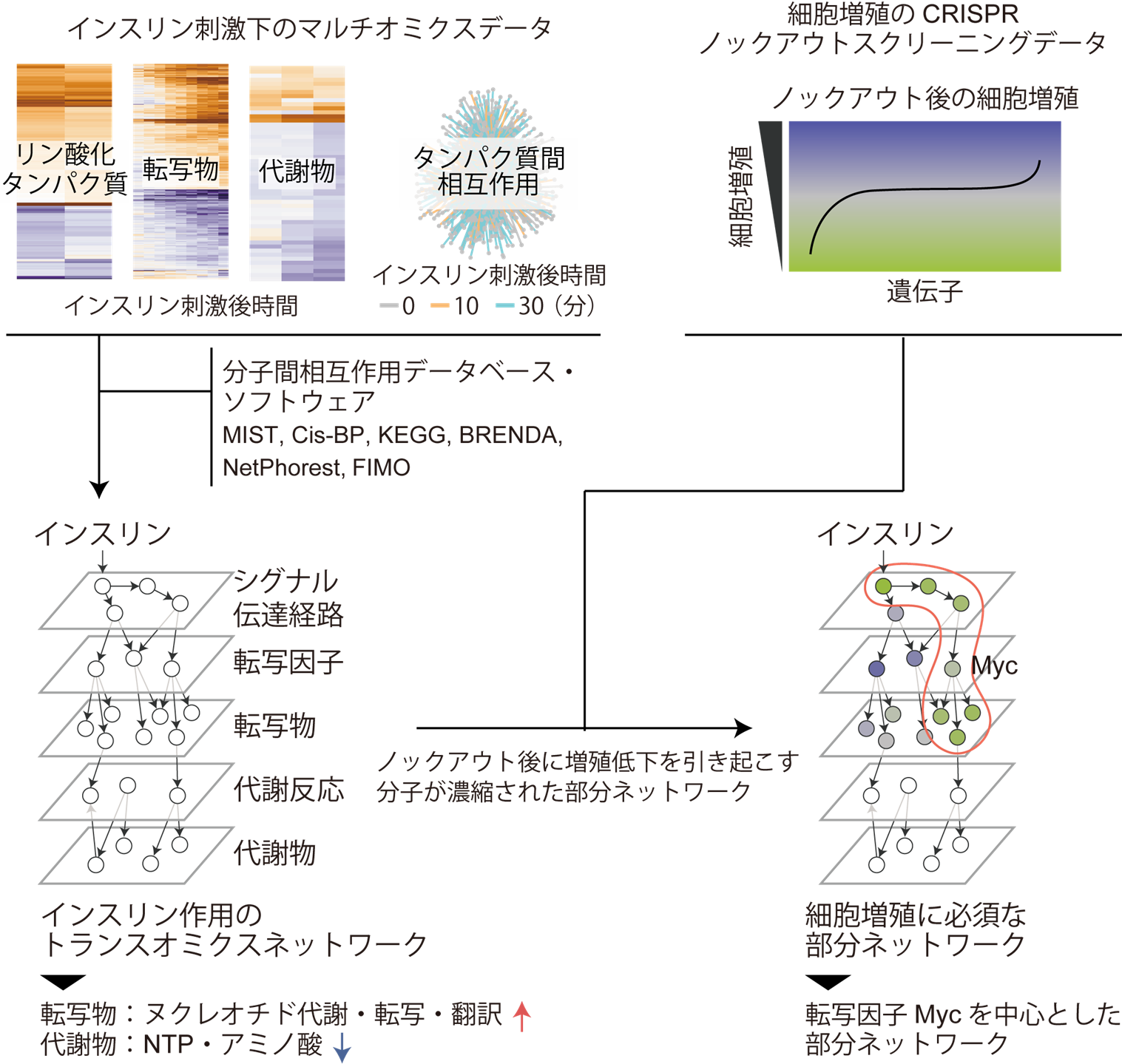
Figure 1: Research Overview
Using insulin-stimulated multi-omics data obtained from Drosophila S2R+ cells (top left) and molecular interaction database software (middle left), a trans-omics network of insulin action was constructed (bottom left). Furthermore, by integrating the transomics network and CRISPR knockout screening data of cell proliferation obtained from Drosophila S2R+ cells (upper right), we estimated the partial network essential for cell proliferation under insulin stimulation (lower right, red frame).
(1) Construction of a trans-omics network of insulin action
First, the research group conducted an integrated analysis of omics data on protein-protein interactions, phosphorylated protein levels, gene expression levels, and metabolite levels obtained from insulin-stimulated Drosophila embryo-derived S2R+ cells, and constructed a trans-omics network that is activated or inactivated under insulin stimulation The data of metabolite abundance were analyzed in this study. Of these, metabolite abundance data were measured in this study, while the other data were measured in previous studies of this group. In constructing the network, we identified groups of molecules that are increased or decreased by insulin stimulation (insulin-responsive molecular groups) and their interactions from each omics data, and estimated the regulatory relationships among the insulin-responsive molecular groups using information from databases on various molecular interactions. In particular, in this study, we extended our previously developed transomics network construction method to incorporate protein-protein interaction data into the network, enabling us to construct a more detailed and comprehensive network than in previous studies.
Analysis of the constructed network revealed a molecular interaction network that functions in insulin-stimulated Drosophila cells. Specifically, a group of 14 transcription factors including the transcription factor Myc was regulated downstream of the insulin signaling pathway (Note 6), and the expression of genes involved in nucleotide metabolism, transcription, and translation was increased through these transcription factors. And at the metabolite level, the amounts of nucleoside triphosphates and amino acids, which are substrates for transcription and translation, were found to be decreased (Figure 1, lower left).
(2) Integrated analysis of CRISPR screening data of trans-omics network of insulin action and cell proliferation
The CRISPR screening data were used to identify the partial network involved in the regulation of proliferation under insulin stimulation (Fig. 1 right). The data can be used to assess the contribution of each molecule in the transomics network to cell proliferation.
The research group evaluated the contribution of each transcription factor, its upstream signaling pathways, and its downstream target genes to cell proliferation in the transomics network (hereinafter referred to as "partial network"). Specifically, since the partial networks involved in the regulation of cell proliferation are expected to contain a large number of molecules that cause a significant reduction in proliferation upon knockout, partial networks that meet these conditions were identified using a method called Gene set enrichment analysis (Note 7). (Note 7).
The analysis revealed that a partial network that promotes expression of nucleotide metabolism and translation-related genes via the transcription factor Myc is involved in the regulation of insulin-stimulated cell proliferation (Fig. 1, bottom right). Myc is known to promote cell growth and proliferation in both Drosophila and mammals, and is also known to be regulated by the insulin signaling pathway, particularly in Drosophila. Thus, in addition to the results showing the high reliability of the developed method, future detailed verification of the causal relationship between the responses of the molecular groups upstream and downstream of Myc and Myc activation inferred in this study is expected to elucidate a novel mechanism of action for insulin action.
Social Significance and Future Plans
However, the large scale of omics data makes them difficult to interpret by simply looking at them, and there has been a need to develop data analysis methods that can extract important phenotype-related findings from omics data. This analysis method is expected to contribute to solving such problems. The framework of this analytical method is applicable to a wide range of species and life phenomena, and it is expected that the developed method will be used to elucidate the mechanisms of action of various life phenomena and diseases in addition to insulin action, which is the subject of this study.
This research is supported by the Japan Science and Technology Agency's Strategic Basic Research Program "Creation of a Quantitative Analysis Platform for Understanding Spatio-Temporal Interactions among Multicellular Organelles," research area entitled "Elucidation of Metabolic Regulation in Multicellular Organelles Using Space-Time Transomics" (project number: JPMJCR2123, PI: Shinya Kuroda). This project was funded by the Grant-in-Aid for Scientific Research on Innovative Areas (Research Area Proposal Type) "Metabolic Adaptation of Type 2 Diabetes Mellitus" (Research Project No.: JP17H06299, JP17H06300; PI: Shinya Kuroda).
Journal
-
Journal name iScience Title of paper Trans-omics analysis of insulin action reveals a cell growth sub-network which co-regulates anabolic processes Author(s) Akira Terakawa, Yanhui Hu, Toshiya Kokaji, Katsuyuki Yugi, Keigo Morita, Satoshi Ohno, Yifei Pan, Yunfan Bai, Andrey A. Parkhitko, Xiaochun Ni, John M. Asara, Martha L. Bulyk, Norbert Perrimriman Asara, Martha L. Bulyk, Norbert Perrimon*, Shinya Kuroda*, and Yunfan Bai DOI Number
Terminology
1 Multi-omics data
The study of biomolecules as a whole in a comprehensive manner is called omics. In addition, multiple types of omics data such as phosphoproteins, transcripts, and metabolites are called multi-omics data. ↑up
Note 2 Mechanism of action
A mechanism that regulates the relevant action through biochemical interactions of biomolecules such as proteins, transcripts, and metabolites. ↑up
Note 3 Omics measurement technology
Technology to comprehensively measure each biomolecular species such as proteins, transcripts, and metabolites. Next-generation sequencers are mainly used to measure transcript levels, while mass spectrometers are mainly used to measure protein levels, protein-protein interactions, and metabolite levels. ↑up
Note 4 Transomics network
A large-scale network spanning signaling pathways, gene expression, and metabolite layers. ↑up
Note 5 CRISPR knockout screening data
Phenotype measurement data when each gene is knocked out using gene knockout technology based on CRISPR-Cas system. In this study, CRISPR screening data for cell proliferation phenotype was used. ↑up
Note 6 Insulin signaling pathway
When insulin binds to insulin receptors on the cell membrane, it regulates gene expression and metabolism through protein phosphorylation and protein-protein interactions. This chain of responses of a group of molecules is called the insulin signaling pathway. ↑up
Note 7 Gene set enrichment analysis
A method to determine whether a particular gene set is biased toward a statistically significantly higher or lower quantitative value, using multiple gene sets and quantitative values associated with each gene as input. In this study, gene sets correspond to groups of genes upstream or downstream of each transcription factor, and quantitative values correspond to CRISPR screening data. ↑


