DATE2022.02.14 #Press Releases
Mechanisms that create inactive regions in plant genomes
Disclaimer: machine translated by DeepL which may contain errors.
Yasuko Fuji, Assistant Professor, Department of Biological Sciences
Tetsuhito Kakutani, Professor, Department of Biological Sciences
Key points of the presentation
- Proliferative sequences that destabilize the genome (Note 1) are subsumed by DNA methylation (Note 2). We have shown that the two types of DNA methylation pathways in plants enhance each other locally.
- On the other hand, we found that there is a genome-wide mechanism that suppresses this mechanism from running amok.
- Mechanisms working in opposite directions locally and throughout the genome may create strongly inactivated regions and prevent genome instability.
Summary of Presentation
Animal and plant genomes contain large amounts of DNA with mobile and proliferative properties, which cause genome instability and diseases such as cancer. DNA methylation is known in plants and vertebrates as a mechanism to inactivate proliferative sequences. However, the mechanism by which only specific DNA sequences in the genome are methylated and inactivated is still largely unknown.
A group led by Assistant Professor Yasuko Fuji and Professor Tetsuhito Kakutani of the Graduate School of Science at The University of Tokyo, using a plant mutant called Arabidopsis thaliana, found a local positive feedback during DNA methylation and a genome-wide negative feedback. Both pathways are thought to prevent genome instability by allowing repression limited to inactivation regions while allowing genes that should be working to work.
Announcement
Transposable elements (TEs), which are proliferative sequences, make up the majority of vertebrate and land plant genomes and contribute greatly to genome evolution. On the other hand, TEs pose a threat to genome stability due to their potential transferability. For this reason, TEs are repressed by multiple epigenetic mechanisms (mechanisms that determine gene on/off information in a form other than DNA sequence and are inherited after cell division), including specific post-translational modifications of histone proteins (Note 3 ), which are major constituent proteins of chromosomes, and DNA methylation.
In plant genomes, repression by DNA methylation is important. Generally, methylation of cytosine is more common in cytosine (CpG) immediately before guanine (G), and methylation of such CpG sequences (mCG) is inherited after cell division by a mechanism called maintenance methylation (Note 4). Cytosine methylation (mCH; H is a base other than G) of sequences other than CpG found in plants is interdependent in that the enzyme catalyzing it is recruited to the methylation-rich region of the 9th lysine residue of the histone H3 protein (H3K9me), and the enzyme catalyzing H3K9me is recruited to the mCH-rich region These modifications are also inherited and enhanced after chromosome division because of the interdependent relationship in which enzymes catalyzing H3K9me are recruited to mCH-rich regions. On the other hand, mCH and mCG are inherited almost independently, and the mechanism by which they both concentrate in TE-rich inactive regions was not understood.
In this study, we found that mCG contributes significantly to the new establishment of mCH and H3K9me in a specific genomic region (Note 5). Surprisingly, this pathway is potent under conditions where mCG is reduced genome-wide. Results with several other mutants also supported the existence of a feedback in which genome-wide DNA methylation negatively regulates mCH induction (Figure 1).
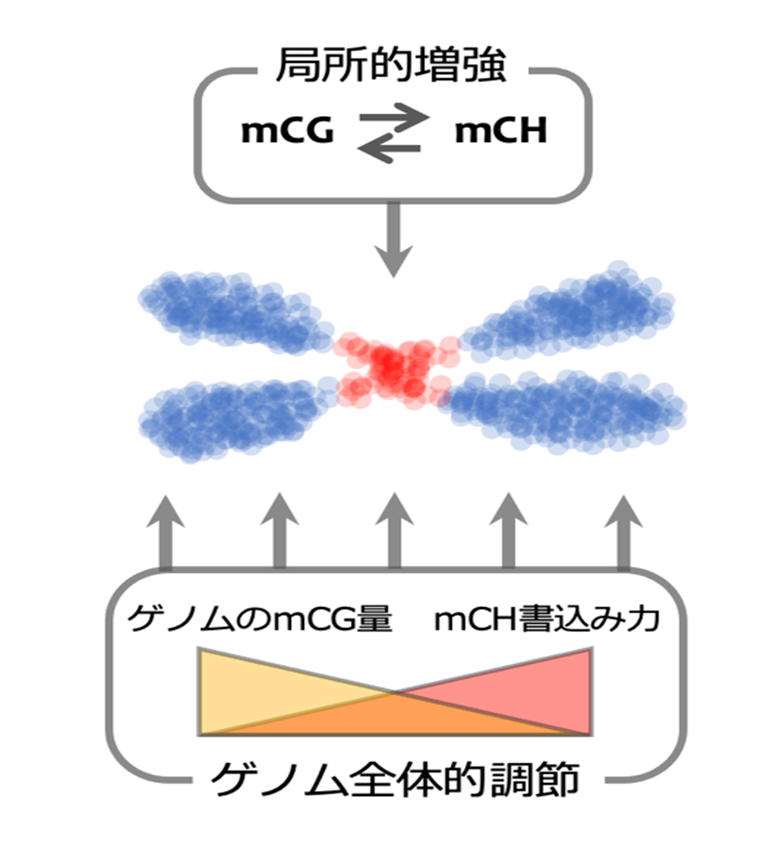
Figure 1: Mechanisms that create inactivation regions in plant genomes.
Plant genomes are broadly classified into transposon-rich inactive regions (red) and gene-rich active regions (blue), each with unique epigenetic modifications. In this study, we found a local positive interaction between two types of DNA methylation (mCG and mCH) and a genome-wide negative interaction. These two interactions may result in the formation of appropriately inactivated regions in the genome.
Since many of the factors mediating DNA methylation and repression by H3K9me are conserved not only in plants but also in animals, it is possible that the mechanisms revealed in this study are conserved in other organisms. In the future, the elucidation of the molecular mechanism of this new pathway is expected to have a major ripple effect.
Journal
-
Journal name Nature Communications Title of paper Local and global crosstalk among heterochromatin marks drives DNA methylome patterning in Arabidopsis Author(s) Taiko Kim To*, Chikae Yamasaki, Shoko Oda, Sayaka Tominaga, Akie Kobayashi, Yoshiaki Tarutani, Tetsuji Kakutani*, Yoshiaki Tarutani, Tetsuji Kakutani*, Yoshiaki Tarutani, Tetsuji Kakutani DOI Number
Terminology
Note 1: Genome
The entire genetic information possessed by a particular organism, or a set of DNA that is its molecular entity. ↑up
Note 2 DNA methylation
In animals and plants, position 5 of cytosine in DNA is sometimes methylated. When such methylation is present near the transcription start site, transcription is often repressed; in addition to methylation of cytosine immediately before G (mCG), methylation immediately before bases other than G (H: A, T, or C) (mCH) is also important in plants. ↑up
Note 3 Post-translational modifications
Some proteins undergo modifications such as methylation and phosphorylation after translation. Such modifications may regulate protein function. ↑up
Note 4 "Inheritance" of DNA methylation
Inheritance of the ON/OFF state of genes after cell division is the basis of epigenetic phenomena, and one of the molecular bases for this is inheritance of DNA methylation. Many plants and animals have an enzyme that methylates the CG sequence of the other strand when there is methylation on one strand of the CG sequence. This enzyme is called a maintenance methyltransferase, and its activity also methylates the CG sequence of the newly synthesized strand during DNA replication. By this mechanism, CG sequences in which both strands are methylated "inherit" the methylation of both strands after DNA replication. On the other hand, mCH in plants is inherited by another pathway via H3K9me. ↑up
Note 5: "Establishment" of DNA methylation
In addition to the above "inheritance," there is a mechanism by which unmethylated cytosines are newly methylated, which is called "establishment" of DNA methylation. In this study, "establishment" activity was detected by introducing a gene that functions in this mutant after loss of DNA methylation under a mutant lacking a factor required for DNA methylation. ↑up


