DATE2021.11.11 #Press Releases
Simultaneous genome editing and fluorescent visualization of four organelles
Disclaimer: machine translated by DeepL which may contain errors.
~Establishment of a new method, "schizon cutter," which opens the door to organelle biology
Naoto Tanaka (2nd year, Master's course, Department of Biological Sciences)
Yuko Mogi (Project Researcher, Department of Biological Sciences)
Tetsuya Higashiyama, Professor, Department of Biological Sciences
Yamato Yoshida, Associate Professor, Department of Biological Sciences
Key points of the presentation
- In the primitive eukaryote unicellular red alga Cizon (Note 1), we have established a new molecular biology tool, the CZON-cutter, which simultaneously realizes fluorescent visualization of four organelles and genome editing (Note 2).
- The CZON-cutter enables rapid analysis of what genes regulate organelles, which are important units that make up cells.
- The schizon cutter has opened the way to a new research field, "organelle biology," which aims to elucidate the true molecular basis of organelle regulation in eukaryotes.
Summary of presentation
Eukaryotic cells contain multiple types of organelles, called organelles, which are surrounded by biological membranes. Each organelle contains specific types of proteins and substrates, which enable various biochemical reactions such as oxidative phosphorylation in mitochondria and photosynthesis in chloroplasts. The existence of such organelles has enabled eukaryotes to realize extremely advanced cellular functions compared to bacteria, which have only a single intracellular compartment, and has led to the emergence of species with various higher-order structures, including humans and plants.
In order for numerous types of organelles to function properly, a large number of genes must work properly. However, many of the molecular mechanisms are still unclear. One of the reasons for the lack of progress in research is that inhibiting the function of genes involved in organelle function often results in the death of the cell itself, making it difficult to analyze the function. Furthermore, with the exception of chloroplasts, which can be seen in visible light, many organelles cannot be seen as they are. Therefore, it has been impossible to detect the presence of organelle abnormalities unless tissue staining is performed to visualize only specific organelles.
To solve the above problem, a research group led by graduate student Naoto Tanaka and Associate Professor Yamato Yoshida at the Graduate School of Science, The University of Tokyo, has developed a new method to analyze organelle function using schizons, which are extremely simple eukaryotes with only one organelle (cell nucleus, mitochondria, chloroplast, and peroxisome) in each cell. We aimed to develop a new molecular biology tool that would enable rapid analysis of organelle function. As a result, we succeeded in establishing the "schizon cutter," a new method to visualize four types of organelles with fluorescent proteins of different colors, and at the same time, to freely modify the sequence of target genes.
With this technology, we were able to establish the basis for elucidating the true molecular mechanism that regulates the organelle, which has yet to be clarified. In the future, it is expected that the schizon cutter will be used to elucidate the principle of organelle formation and to advance our understanding of how eukaryotic organisms are born.
Contents of presentation
One of the major differences between eukaryotic and bacterial cell structures is the presence of multiple types of organelles surrounded by biological membranes. However, the molecular mechanisms that control the division, proliferation, and various functions of these organelles in eukaryotes have not yet been fully elucidated, leaving this as an important research issue.
In this study, a research group led by graduate student Naoto Tanaka and Associate Professor Yamato Yoshida at the Graduate School of Science, The University of Tokyo, attempted to develop a new molecular biological tool to analyze the molecular mechanisms by which eukaryotic organisms regulate organelles. In this study, they used the unicellular red alga Schizon, which retains many primitive features. The schizon has an extremely simple cell structure, with only one cell nucleus, one mitochondrion, one chloroplast, and one membrane organelle such as a peroxisome per cell. The genome DNA has been decoded, and it is clear that only 4775 genes are encoded, the smallest number among eukaryotes, making it the simplest eukaryote in terms of genome structure. In addition, recent estimates of the evolutionary rate based on molecular clock analysis, which utilizes the rate of introduction of mutations into gene sequences, suggest that the schizon lineage was born about 1.6 billion years ago in the middle of the Proterozoic Era, making it a "living fossil" that still retains the characteristics of primitive eukaryotes. Because schizon is such an extremely important organism for studying the basic principles of eukaryotes, the research group has attempted to develop a new method that will strongly encourage further research progress.
The research group constructed the schizon YMT1 strain, which incorporates Cas9, a fusion of the fluorescent protein Venus and nuclear localization signals, into the genomic DNA of schizons (Figure 1).
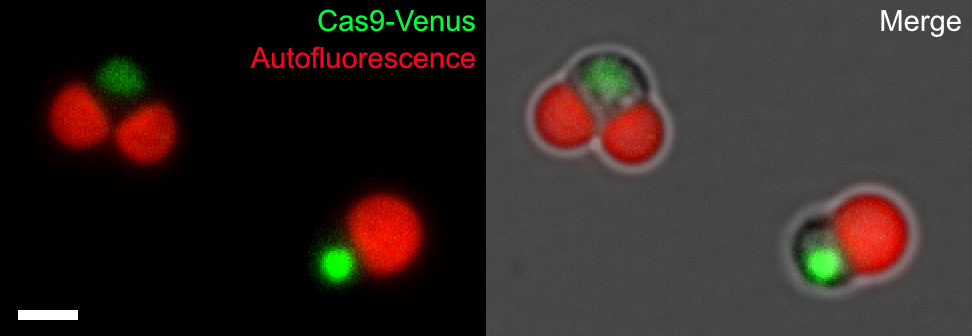
Figure 1: (Left) Schizon YMT1 incorporating NLS-Cas9-NLS-Venus (Cas9-Venus)
Cas9-Venus (yellow-green) fused with nuclear transfer signal (NLS) localizes to the cell nucleus, chloroplasts are detected by chlorophyll autofluorescence (red). Scale bar is 2 μm.
Next, a reporter gene cassette encoding an orange fluorescent protein mScarlet fused with a sequence encoding a single guide RNA (sgRNA) to cleave the target genomic DNA sequence and a mitochondrial transition signal was introduced into cells of this schizon YMT1 strain. By mixing DNA fragments with arbitrary sequences in this process, it was possible to visualize cell nuclei, mitochondria, and chloroplasts while freely editing the genome sequence (Figure 2). We also succeeded in inserting a gene cassette encoding mCerulean3, a blue fluorescent protein fused with a peroxisome migration signal, into the truncated genomic DNA, thus enabling visualization of cell nuclei (Venus, yellow-green), mitochondria (mScarlet, orange), peroxisomes ( This has enabled fluorescent observation of four types of organelles: nucleus (Venus, yellow-green), mitochondria (mCerulean3, blue), peroxisomes (mCerulean3, blue), and chloroplasts (chlorophyll autofluorescence, red) (Figure 3).
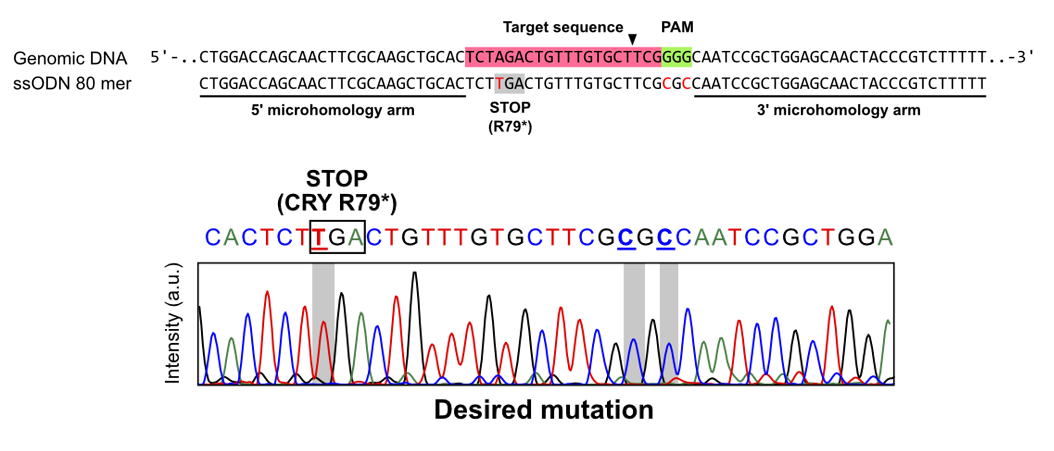
Figure 2: Genomic DNA sequences using single-stranded oligo DNA
The target sequence targeted by sgRNA (upper row: red highlight) was cleaved and an attempt was made to modify the genomic DNA sequence using an 80-base single-stranded oligo DNA as a template, and the Sanger sequencing method confirmed that the target sequence was actually edited (lower row: gray highlight).
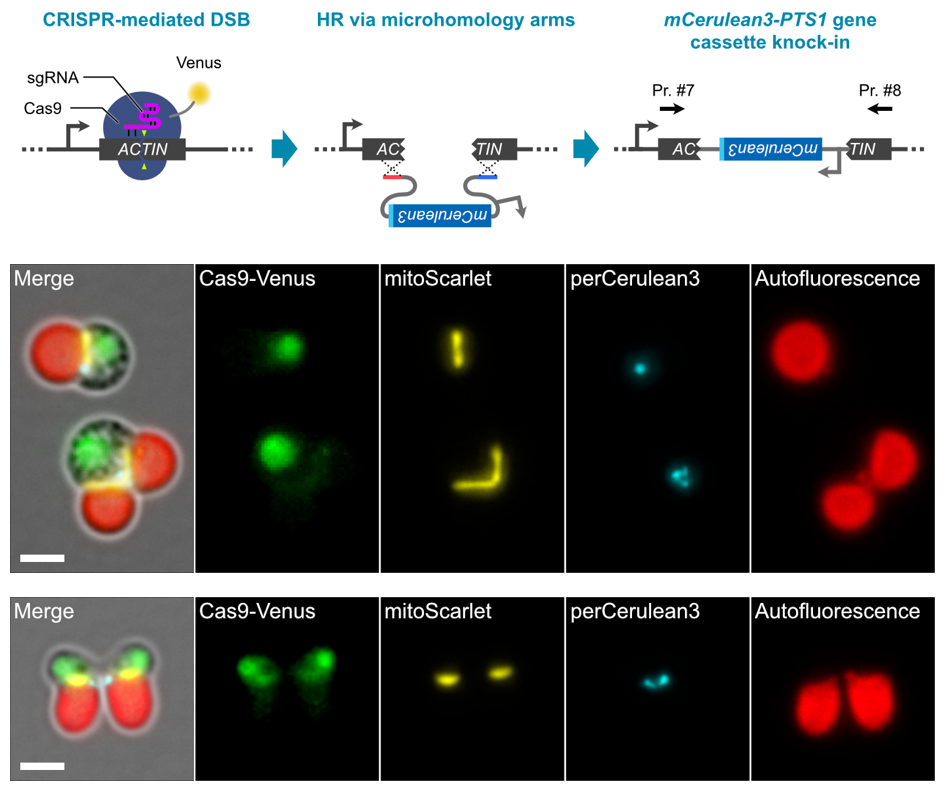
Figure 3: Knockout of ACTIN gene by gene cassette knock-in
(Upper row) The genomic region of the ACTIN gene was truncated and the perCerulean3 gene cassette, in which a peroxisomal transfer signal was added to mCerulean3, was knocked in. (Middle) The resulting transformant line can be live-imaged with fluorescence of four organelles: cell nucleus, mitochondria, peroxisomes, and chloroplasts. (Lower row) Normal cell division occurs despite knockout of the ACTIN gene, which is normally essential for cell division, suggesting that an unknown cell division mechanism may be at work. Scale bar is 2 μm.
The CRISPR-Cas9-based genome editing and organelle fluorescence imaging method "Schizon Cutter" established in this study can easily and efficiently modify any gene, no matter what the target gene is, by simply changing a 20-nucleotide sgRNA that specifies the genomic region to be modified. The new method enables easy and efficient genetic modification and organelle fluorescence imaging of any target gene through a completely systematic process. This method has enabled genome-wide high-throughput analysis, which was not possible before, and we will now develop organelle biology, which aims to elucidate the true molecular basis of organelle regulation in eukaryotes.
This research was supported by JST PRESTO "Elucidation of the molecular mechanisms of the organelle fission ring" (project number: JPMJPR20EE, PI: Yamato Yoshida), the Human Frontier Science Program Career Development Award "Decoding the molecular mechanisms and kinetics of the Decoding the molecular mechanisms and kinetics of the plastid- and mitochondrial-division machinery" (project number: CDA00049/2018-C, PI: Yamato Yoshida), and "Decoding the molecular mechanisms and kinetics of the plastid- and mitochondrial-division machinery" (project number: CDA00049/2018-C, PI: Yamato Yoshida). The Institute for Fermentation Research, Japan, Grant-in-Aid for Scientific Research, "Development of a New Paradigm and Elucidation of Molecular Mechanisms of Organelle and Cell Division and Growth Mechanisms Based on Genome-Wide Central Dogma Analysis" (Project Leader: Yamato Yoshida), Grant-in-Aid for Scientific Research, "Morphological Structure and Molecular Mechanism of Mitochondrial Division Apparatus" (Project Number: JP18K06325, PI: Yamato Yoshida) (Project number: L-2020-2-008, PI: Yamato Yoshida), Sumitomo Foundation Basic Science Research Grant, "Elucidation of the Central Dogma Regulating Mitochondrial and Chloroplast Division and Proliferation" (Project number: 180705, PI: Yamato Yoshida), and the Grant-in-Aid for Scientific Research on Innovative Areas "Principles for the Emergence of New Plant Species: Through Elucidation of Molecular Mechanisms of Key and Keyhole of Reproductive Processes (Project Leader: Tetsuya Higashiyama), and CREST "Higher-order Membrane Traffic Dynamics Driving Chemotaxis" (Project Leader: Tetsuya Higashiyama, JPMJCR20E5).
Journal
-
Journal name Journal of Cell ScienceTitle of paper CZON-cutter: a CRISPR-Cas9 system for multiplexed organelle imaging in a simple unicellular algaAuthor(s) Naoto Tanaka, Yuko Mogi, Takayuki Fujiwara, Kannosuke Yabe, Yukiho Toyama, Tetsuya Higashiyama, and Yamato Yoshida* (author)DOI Number
Terminology
Note 1: The unicellular red alga Cyanidioschyzon
Cyanidioschyzon merolae has an extremely simple structure, with only one major organelle per cell (nucleus, mitochondria, chloroplast). The 16.5 Mbp genome encodes only 4775 genes, the smallest among eukaryotes, and contains only 14 genes including introns. Cell division can be synchronized by culturing the cells under light-dark cycles. It is of particular interest as an important experimental organism for studying basic eukaryotic cell functions and organelles. ↑ upNote 2 Genome editing
A technique to modify any genomic DNA sequence using nucleases such as ZFN, TALEN, and Cas9. In this study, CRISPR/Cas9 genome editing using Cas9 protein isolated from Streptococcus pyogenes is used. Cas9 protein is induced and the target DNA sequence is cleaved. The genomic DNA sequence can then be altered to the sequence of the template DNA by coexisting with a template DNA fragment as the cleaved DNA is repaired. ↑ up


